
Candidatus Thiomargarita nelsonii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria<
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
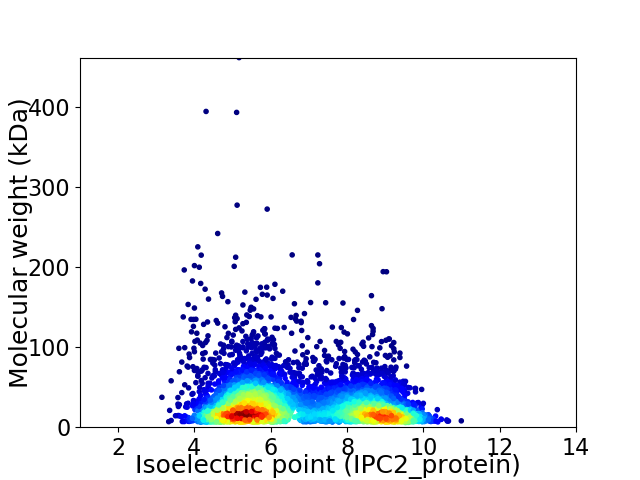
Virtual 2D-PAGE plot for 5897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4E0QWB0|A0A4E0QWB0_9GAMM Uncharacterized protein OS=Candidatus Thiomargarita nelsonii OX=1003181 GN=PN36_09985 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84SNIKK6 pKa = 10.29KK7 pKa = 8.37QTLEE11 pKa = 3.88HH12 pKa = 7.04AICGPQTKK20 pKa = 9.11PWRR23 pKa = 11.84SSLASSLNSLCPSSNWRR40 pKa = 11.84GVVFVAFIVGIMAIFASTQVQAVPIPGVSEE70 pKa = 4.14YY71 pKa = 10.75FAGTVTDD78 pKa = 3.7QNGDD82 pKa = 4.14PISDD86 pKa = 3.25ATVQINEE93 pKa = 4.17VADD96 pKa = 3.4NTEE99 pKa = 3.91RR100 pKa = 11.84DD101 pKa = 3.68GSFGFHH107 pKa = 6.58VEE109 pKa = 5.52RR110 pKa = 11.84DD111 pKa = 3.73DD112 pKa = 3.64QDD114 pKa = 3.52QYY116 pKa = 11.42IINVTKK122 pKa = 10.42RR123 pKa = 11.84GYY125 pKa = 11.18ALVSQIEE132 pKa = 4.16HH133 pKa = 6.72APNVSLRR140 pKa = 11.84FTLKK144 pKa = 9.49QAEE147 pKa = 4.38VFIIDD152 pKa = 3.89PSQTSKK158 pKa = 11.28VEE160 pKa = 4.02DD161 pKa = 3.45SRR163 pKa = 11.84GTRR166 pKa = 11.84IMLPPNSLVDD176 pKa = 3.81SDD178 pKa = 4.96GNPPDD183 pKa = 5.17GEE185 pKa = 4.42VQMGLYY191 pKa = 9.85TYY193 pKa = 11.14DD194 pKa = 3.86LANEE198 pKa = 4.17SMPGDD203 pKa = 3.3MGAINSQGEE212 pKa = 4.2PGAMLSAGAFYY223 pKa = 11.26AEE225 pKa = 4.34FTDD228 pKa = 4.82DD229 pKa = 3.41SGRR232 pKa = 11.84MYY234 pKa = 11.21DD235 pKa = 3.77LASDD239 pKa = 4.03MVAEE243 pKa = 4.38ISIPAINSEE252 pKa = 4.28EE253 pKa = 4.33TVGLWDD259 pKa = 3.72YY260 pKa = 11.67NKK262 pKa = 9.01EE263 pKa = 4.11TGKK266 pKa = 10.03WMEE269 pKa = 5.29DD270 pKa = 3.21GSASLINGRR279 pKa = 11.84FEE281 pKa = 4.04GTVKK285 pKa = 10.53HH286 pKa = 5.55FSVWNFDD293 pKa = 3.31LFYY296 pKa = 11.42NGVACVKK303 pKa = 10.81LEE305 pKa = 3.94IAQAFYY311 pKa = 9.4DD312 pKa = 4.05TYY314 pKa = 11.5KK315 pKa = 11.03DD316 pKa = 3.44GAGVLKK322 pKa = 10.27IQAVVTTPGFSPITDD337 pKa = 3.64VLSMDD342 pKa = 4.25EE343 pKa = 4.21YY344 pKa = 11.49NYY346 pKa = 10.81DD347 pKa = 4.47DD348 pKa = 5.75PYY350 pKa = 11.92ALYY353 pKa = 10.32NLPPNATVDD362 pKa = 4.27FYY364 pKa = 11.89VPPPFSLPPYY374 pKa = 9.53ASVSTGAAWGGTGFPPHH391 pKa = 7.27PYY393 pKa = 9.69DD394 pKa = 3.34VCNGSMVLNFPFSNDD409 pKa = 3.17VIIDD413 pKa = 4.12FGTQNGILVWMNNSAWVQLHH433 pKa = 6.21PLSPDD438 pKa = 3.08SMVTGDD444 pKa = 3.94MDD446 pKa = 5.5GNGQDD451 pKa = 4.85DD452 pKa = 4.43VIIDD456 pKa = 4.14FGSPHH461 pKa = 7.41GILVWMNNSAWVQLHH476 pKa = 6.19PLSPSSMITGDD487 pKa = 3.17MDD489 pKa = 5.08GNGLDD494 pKa = 4.55DD495 pKa = 5.19VIIDD499 pKa = 3.71FPGYY503 pKa = 9.94GIWVMMNNSTWVQLHH518 pKa = 5.78SLSPSSMTTGDD529 pKa = 3.18MDD531 pKa = 5.13GNGLDD536 pKa = 4.55DD537 pKa = 5.19VIIDD541 pKa = 3.71FPGYY545 pKa = 9.94GIWVMMNNSTWVQLHH560 pKa = 5.78SLSPSSMTTGDD571 pKa = 3.18MDD573 pKa = 5.13GNGLDD578 pKa = 4.55DD579 pKa = 5.19VIIDD583 pKa = 3.71FPGYY587 pKa = 9.94GIWVMMNNSTWVQLHH602 pKa = 5.78SLSPSSMTTGDD613 pKa = 3.18MDD615 pKa = 5.13GNGLDD620 pKa = 4.55DD621 pKa = 5.19VIIDD625 pKa = 3.71FPGYY629 pKa = 9.94GIWVMMNNSTWVNLHH644 pKa = 5.93SLSPSSMTTGDD655 pKa = 4.14LDD657 pKa = 3.97NNAQADD663 pKa = 4.13VIIDD667 pKa = 3.81FGPGVGILVMMNNSTWVTLHH687 pKa = 6.53SNSADD692 pKa = 3.3SMVTGNIDD700 pKa = 3.33GLPP703 pKa = 3.44
MM1 pKa = 7.9RR2 pKa = 11.84SNIKK6 pKa = 10.29KK7 pKa = 8.37QTLEE11 pKa = 3.88HH12 pKa = 7.04AICGPQTKK20 pKa = 9.11PWRR23 pKa = 11.84SSLASSLNSLCPSSNWRR40 pKa = 11.84GVVFVAFIVGIMAIFASTQVQAVPIPGVSEE70 pKa = 4.14YY71 pKa = 10.75FAGTVTDD78 pKa = 3.7QNGDD82 pKa = 4.14PISDD86 pKa = 3.25ATVQINEE93 pKa = 4.17VADD96 pKa = 3.4NTEE99 pKa = 3.91RR100 pKa = 11.84DD101 pKa = 3.68GSFGFHH107 pKa = 6.58VEE109 pKa = 5.52RR110 pKa = 11.84DD111 pKa = 3.73DD112 pKa = 3.64QDD114 pKa = 3.52QYY116 pKa = 11.42IINVTKK122 pKa = 10.42RR123 pKa = 11.84GYY125 pKa = 11.18ALVSQIEE132 pKa = 4.16HH133 pKa = 6.72APNVSLRR140 pKa = 11.84FTLKK144 pKa = 9.49QAEE147 pKa = 4.38VFIIDD152 pKa = 3.89PSQTSKK158 pKa = 11.28VEE160 pKa = 4.02DD161 pKa = 3.45SRR163 pKa = 11.84GTRR166 pKa = 11.84IMLPPNSLVDD176 pKa = 3.81SDD178 pKa = 4.96GNPPDD183 pKa = 5.17GEE185 pKa = 4.42VQMGLYY191 pKa = 9.85TYY193 pKa = 11.14DD194 pKa = 3.86LANEE198 pKa = 4.17SMPGDD203 pKa = 3.3MGAINSQGEE212 pKa = 4.2PGAMLSAGAFYY223 pKa = 11.26AEE225 pKa = 4.34FTDD228 pKa = 4.82DD229 pKa = 3.41SGRR232 pKa = 11.84MYY234 pKa = 11.21DD235 pKa = 3.77LASDD239 pKa = 4.03MVAEE243 pKa = 4.38ISIPAINSEE252 pKa = 4.28EE253 pKa = 4.33TVGLWDD259 pKa = 3.72YY260 pKa = 11.67NKK262 pKa = 9.01EE263 pKa = 4.11TGKK266 pKa = 10.03WMEE269 pKa = 5.29DD270 pKa = 3.21GSASLINGRR279 pKa = 11.84FEE281 pKa = 4.04GTVKK285 pKa = 10.53HH286 pKa = 5.55FSVWNFDD293 pKa = 3.31LFYY296 pKa = 11.42NGVACVKK303 pKa = 10.81LEE305 pKa = 3.94IAQAFYY311 pKa = 9.4DD312 pKa = 4.05TYY314 pKa = 11.5KK315 pKa = 11.03DD316 pKa = 3.44GAGVLKK322 pKa = 10.27IQAVVTTPGFSPITDD337 pKa = 3.64VLSMDD342 pKa = 4.25EE343 pKa = 4.21YY344 pKa = 11.49NYY346 pKa = 10.81DD347 pKa = 4.47DD348 pKa = 5.75PYY350 pKa = 11.92ALYY353 pKa = 10.32NLPPNATVDD362 pKa = 4.27FYY364 pKa = 11.89VPPPFSLPPYY374 pKa = 9.53ASVSTGAAWGGTGFPPHH391 pKa = 7.27PYY393 pKa = 9.69DD394 pKa = 3.34VCNGSMVLNFPFSNDD409 pKa = 3.17VIIDD413 pKa = 4.12FGTQNGILVWMNNSAWVQLHH433 pKa = 6.21PLSPDD438 pKa = 3.08SMVTGDD444 pKa = 3.94MDD446 pKa = 5.5GNGQDD451 pKa = 4.85DD452 pKa = 4.43VIIDD456 pKa = 4.14FGSPHH461 pKa = 7.41GILVWMNNSAWVQLHH476 pKa = 6.19PLSPSSMITGDD487 pKa = 3.17MDD489 pKa = 5.08GNGLDD494 pKa = 4.55DD495 pKa = 5.19VIIDD499 pKa = 3.71FPGYY503 pKa = 9.94GIWVMMNNSTWVQLHH518 pKa = 5.78SLSPSSMTTGDD529 pKa = 3.18MDD531 pKa = 5.13GNGLDD536 pKa = 4.55DD537 pKa = 5.19VIIDD541 pKa = 3.71FPGYY545 pKa = 9.94GIWVMMNNSTWVQLHH560 pKa = 5.78SLSPSSMTTGDD571 pKa = 3.18MDD573 pKa = 5.13GNGLDD578 pKa = 4.55DD579 pKa = 5.19VIIDD583 pKa = 3.71FPGYY587 pKa = 9.94GIWVMMNNSTWVQLHH602 pKa = 5.78SLSPSSMTTGDD613 pKa = 3.18MDD615 pKa = 5.13GNGLDD620 pKa = 4.55DD621 pKa = 5.19VIIDD625 pKa = 3.71FPGYY629 pKa = 9.94GIWVMMNNSTWVNLHH644 pKa = 5.93SLSPSSMTTGDD655 pKa = 4.14LDD657 pKa = 3.97NNAQADD663 pKa = 4.13VIIDD667 pKa = 3.81FGPGVGILVMMNNSTWVTLHH687 pKa = 6.53SNSADD692 pKa = 3.3SMVTGNIDD700 pKa = 3.33GLPP703 pKa = 3.44
Molecular weight: 76.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4E0QLU1|A0A4E0QLU1_9GAMM Uncharacterized protein OS=Candidatus Thiomargarita nelsonii OX=1003181 GN=PN36_35000 PE=4 SV=1
MM1 pKa = 7.32HH2 pKa = 7.64RR3 pKa = 11.84ALSLKK8 pKa = 10.29NFSLFLLRR16 pKa = 11.84GLLTGNTLRR25 pKa = 11.84LAQTIIWRR33 pKa = 11.84TKK35 pKa = 9.33KK36 pKa = 10.54KK37 pKa = 10.48SIRR40 pKa = 11.84PQINMKK46 pKa = 10.34RR47 pKa = 11.84LIKK50 pKa = 10.23PVVKK54 pKa = 9.37PVLMIQVLAVFQQMM68 pKa = 3.65
MM1 pKa = 7.32HH2 pKa = 7.64RR3 pKa = 11.84ALSLKK8 pKa = 10.29NFSLFLLRR16 pKa = 11.84GLLTGNTLRR25 pKa = 11.84LAQTIIWRR33 pKa = 11.84TKK35 pKa = 9.33KK36 pKa = 10.54KK37 pKa = 10.48SIRR40 pKa = 11.84PQINMKK46 pKa = 10.34RR47 pKa = 11.84LIKK50 pKa = 10.23PVVKK54 pKa = 9.37PVLMIQVLAVFQQMM68 pKa = 3.65
Molecular weight: 7.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1621432 |
51 |
4061 |
275.0 |
31.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.362 ± 0.038 | 1.07 ± 0.012 |
5.272 ± 0.026 | 6.474 ± 0.036 |
4.366 ± 0.024 | 6.272 ± 0.037 |
2.257 ± 0.017 | 6.997 ± 0.032 |
6.242 ± 0.041 | 10.677 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.017 | 4.564 ± 0.027 |
4.199 ± 0.022 | 4.701 ± 0.033 |
4.898 ± 0.026 | 6.066 ± 0.029 |
5.417 ± 0.033 | 6.051 ± 0.028 |
1.471 ± 0.014 | 3.447 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
