
Wuhan cricket virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
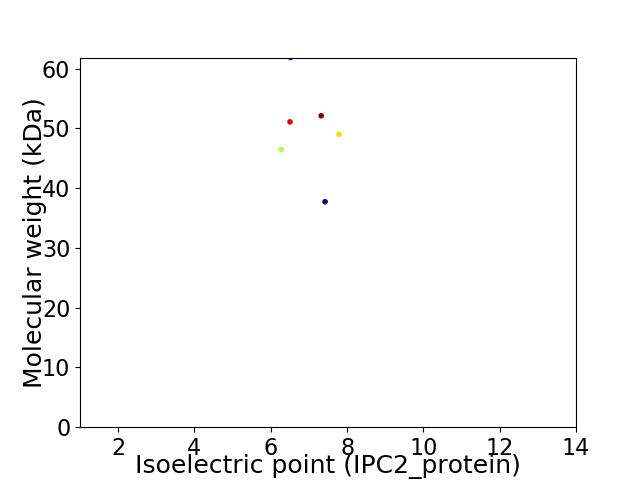
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLR6|A0A1L3KLR6_9VIRU Uncharacterized protein OS=Wuhan cricket virus 2 OX=1923697 PE=4 SV=1
MM1 pKa = 7.79EE2 pKa = 6.0DD3 pKa = 3.65LPRR6 pKa = 11.84VAPEE10 pKa = 3.64DD11 pKa = 3.94SIFARR16 pKa = 11.84YY17 pKa = 8.63SWEE20 pKa = 4.03FNPDD24 pKa = 3.05SLVPASSRR32 pKa = 11.84QTSRR36 pKa = 11.84RR37 pKa = 11.84LDD39 pKa = 3.77LNPNVQTTPQQPASPPHH56 pKa = 7.03HH57 pKa = 6.76GMRR60 pKa = 11.84TIQEE64 pKa = 3.88WASLVRR70 pKa = 11.84EE71 pKa = 4.84HH72 pKa = 6.07IQKK75 pKa = 10.35RR76 pKa = 11.84MARR79 pKa = 11.84EE80 pKa = 3.68PYY82 pKa = 7.96MTIGEE87 pKa = 4.18QMKK90 pKa = 10.47LLSLEE95 pKa = 4.52DD96 pKa = 3.52FHH98 pKa = 9.38IEE100 pKa = 3.88PSVPAATTLHH110 pKa = 6.68INLPDD115 pKa = 3.57NTEE118 pKa = 3.64EE119 pKa = 4.76AIPVPSDD126 pKa = 3.3LTIDD130 pKa = 3.42TANAKK135 pKa = 9.5LHH137 pKa = 5.39QAEE140 pKa = 4.18QPVIRR145 pKa = 11.84AIGSVAATNVIAEE158 pKa = 4.41TEE160 pKa = 4.09RR161 pKa = 11.84VITNHH166 pKa = 5.26NRR168 pKa = 11.84EE169 pKa = 3.97IMSDD173 pKa = 3.74DD174 pKa = 3.48LHH176 pKa = 7.21KK177 pKa = 10.84VWVMNHH183 pKa = 4.83FVRR186 pKa = 11.84RR187 pKa = 11.84IPLSHH192 pKa = 6.31EE193 pKa = 3.87THH195 pKa = 6.1HH196 pKa = 6.63QRR198 pKa = 11.84GLPLPTFSSSPEE210 pKa = 3.62AKK212 pKa = 9.5IARR215 pKa = 11.84TSTPLLDD222 pKa = 3.88SRR224 pKa = 11.84PPSISPSSWSASDD237 pKa = 3.46IASDD241 pKa = 5.22SPPQLPRR248 pKa = 11.84KK249 pKa = 8.8LAPHH253 pKa = 6.89LPAAPQKK260 pKa = 10.18EE261 pKa = 4.04MRR263 pKa = 11.84YY264 pKa = 9.57LLSATLTSSAPEE276 pKa = 3.45ALFFTEE282 pKa = 4.58EE283 pKa = 3.83ASKK286 pKa = 11.29NPIVGSTLVKK296 pKa = 8.76QTYY299 pKa = 10.5GLLHH303 pKa = 7.2DD304 pKa = 4.81WLAAMSFKK312 pKa = 10.38MRR314 pKa = 11.84TSQVFKK320 pKa = 10.92LRR322 pKa = 11.84NTLACIHH329 pKa = 6.31CHH331 pKa = 5.21QSVNMAYY338 pKa = 10.33NQVTHH343 pKa = 5.68NHH345 pKa = 5.78RR346 pKa = 11.84CVAYY350 pKa = 10.13ALLASFTDD358 pKa = 3.87TVSCQDD364 pKa = 3.86GLHH367 pKa = 6.57HH368 pKa = 7.04LPQYY372 pKa = 10.12SAPLYY377 pKa = 9.59YY378 pKa = 10.33RR379 pKa = 11.84LVRR382 pKa = 11.84NLRR385 pKa = 11.84ISHH388 pKa = 7.13PLRR391 pKa = 11.84QLFSCHH397 pKa = 6.79DD398 pKa = 3.71GVFTPLQTTWLVDD411 pKa = 3.5DD412 pKa = 4.25TT413 pKa = 4.69
MM1 pKa = 7.79EE2 pKa = 6.0DD3 pKa = 3.65LPRR6 pKa = 11.84VAPEE10 pKa = 3.64DD11 pKa = 3.94SIFARR16 pKa = 11.84YY17 pKa = 8.63SWEE20 pKa = 4.03FNPDD24 pKa = 3.05SLVPASSRR32 pKa = 11.84QTSRR36 pKa = 11.84RR37 pKa = 11.84LDD39 pKa = 3.77LNPNVQTTPQQPASPPHH56 pKa = 7.03HH57 pKa = 6.76GMRR60 pKa = 11.84TIQEE64 pKa = 3.88WASLVRR70 pKa = 11.84EE71 pKa = 4.84HH72 pKa = 6.07IQKK75 pKa = 10.35RR76 pKa = 11.84MARR79 pKa = 11.84EE80 pKa = 3.68PYY82 pKa = 7.96MTIGEE87 pKa = 4.18QMKK90 pKa = 10.47LLSLEE95 pKa = 4.52DD96 pKa = 3.52FHH98 pKa = 9.38IEE100 pKa = 3.88PSVPAATTLHH110 pKa = 6.68INLPDD115 pKa = 3.57NTEE118 pKa = 3.64EE119 pKa = 4.76AIPVPSDD126 pKa = 3.3LTIDD130 pKa = 3.42TANAKK135 pKa = 9.5LHH137 pKa = 5.39QAEE140 pKa = 4.18QPVIRR145 pKa = 11.84AIGSVAATNVIAEE158 pKa = 4.41TEE160 pKa = 4.09RR161 pKa = 11.84VITNHH166 pKa = 5.26NRR168 pKa = 11.84EE169 pKa = 3.97IMSDD173 pKa = 3.74DD174 pKa = 3.48LHH176 pKa = 7.21KK177 pKa = 10.84VWVMNHH183 pKa = 4.83FVRR186 pKa = 11.84RR187 pKa = 11.84IPLSHH192 pKa = 6.31EE193 pKa = 3.87THH195 pKa = 6.1HH196 pKa = 6.63QRR198 pKa = 11.84GLPLPTFSSSPEE210 pKa = 3.62AKK212 pKa = 9.5IARR215 pKa = 11.84TSTPLLDD222 pKa = 3.88SRR224 pKa = 11.84PPSISPSSWSASDD237 pKa = 3.46IASDD241 pKa = 5.22SPPQLPRR248 pKa = 11.84KK249 pKa = 8.8LAPHH253 pKa = 6.89LPAAPQKK260 pKa = 10.18EE261 pKa = 4.04MRR263 pKa = 11.84YY264 pKa = 9.57LLSATLTSSAPEE276 pKa = 3.45ALFFTEE282 pKa = 4.58EE283 pKa = 3.83ASKK286 pKa = 11.29NPIVGSTLVKK296 pKa = 8.76QTYY299 pKa = 10.5GLLHH303 pKa = 7.2DD304 pKa = 4.81WLAAMSFKK312 pKa = 10.38MRR314 pKa = 11.84TSQVFKK320 pKa = 10.92LRR322 pKa = 11.84NTLACIHH329 pKa = 6.31CHH331 pKa = 5.21QSVNMAYY338 pKa = 10.33NQVTHH343 pKa = 5.68NHH345 pKa = 5.78RR346 pKa = 11.84CVAYY350 pKa = 10.13ALLASFTDD358 pKa = 3.87TVSCQDD364 pKa = 3.86GLHH367 pKa = 6.57HH368 pKa = 7.04LPQYY372 pKa = 10.12SAPLYY377 pKa = 9.59YY378 pKa = 10.33RR379 pKa = 11.84LVRR382 pKa = 11.84NLRR385 pKa = 11.84ISHH388 pKa = 7.13PLRR391 pKa = 11.84QLFSCHH397 pKa = 6.79DD398 pKa = 3.71GVFTPLQTTWLVDD411 pKa = 3.5DD412 pKa = 4.25TT413 pKa = 4.69
Molecular weight: 46.46 kDa
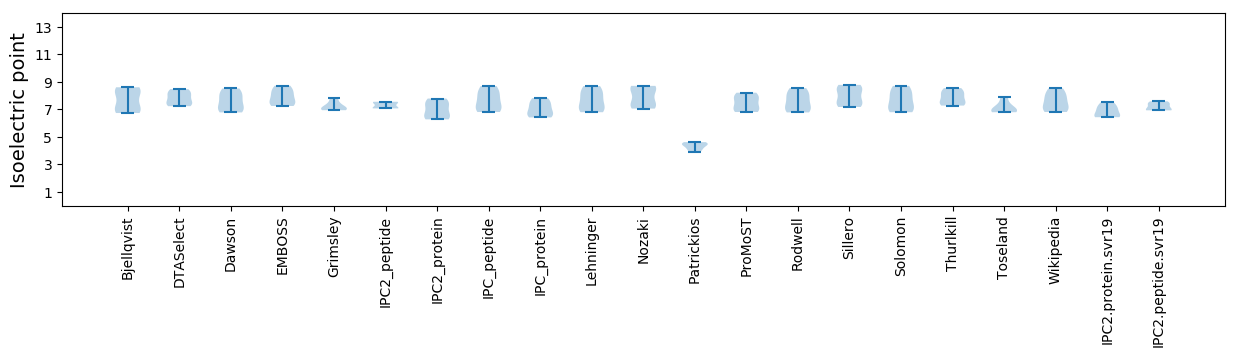
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLY0|A0A1L3KLY0_9VIRU Putative capsid protein OS=Wuhan cricket virus 2 OX=1923697 PE=4 SV=1
MM1 pKa = 8.17DD2 pKa = 5.04PRR4 pKa = 11.84PYY6 pKa = 10.33LGKK9 pKa = 9.35PANQKK14 pKa = 9.87NPRR17 pKa = 11.84AKK19 pKa = 10.08NLHH22 pKa = 5.37VPRR25 pKa = 11.84APRR28 pKa = 11.84EE29 pKa = 4.02APAAMEE35 pKa = 4.47DD36 pKa = 4.08TPPAPSKK43 pKa = 10.43QASKK47 pKa = 10.95PKK49 pKa = 9.76MPSTLPPPKK58 pKa = 9.72HH59 pKa = 6.48ASNPPRR65 pKa = 11.84KK66 pKa = 9.1EE67 pKa = 3.85DD68 pKa = 3.75FVEE71 pKa = 4.56KK72 pKa = 10.81SPLSDD77 pKa = 5.36AIFQEE82 pKa = 4.29DD83 pKa = 3.85MAVRR87 pKa = 11.84EE88 pKa = 4.37VNLTTRR94 pKa = 11.84QNVSLSAVNEE104 pKa = 4.09LSRR107 pKa = 11.84ATYY110 pKa = 9.94QQMLITDD117 pKa = 4.41PNLAKK122 pKa = 10.11QWTPEE127 pKa = 3.6VHH129 pKa = 7.18DD130 pKa = 4.5YY131 pKa = 11.36YY132 pKa = 10.12VTAMTWLRR140 pKa = 11.84IVALKK145 pKa = 10.63ASSGQDD151 pKa = 3.1LTPAEE156 pKa = 4.26EE157 pKa = 4.45TLLSMSATHH166 pKa = 6.49SFNLTEE172 pKa = 5.19PIRR175 pKa = 11.84LYY177 pKa = 11.09LSGVGVAVTKK187 pKa = 10.66NGQHH191 pKa = 7.28LYY193 pKa = 10.33PSFPPLPTEE202 pKa = 4.24VVDD205 pKa = 6.1GIPGLYY211 pKa = 10.38GPIGEE216 pKa = 4.56ATHH219 pKa = 6.32NLYY222 pKa = 10.9EE223 pKa = 4.73EE224 pKa = 4.55IPALGVSIGMIRR236 pKa = 11.84ASLNPVRR243 pKa = 11.84PIPQWVPPILPVNTNANPNLLGFRR267 pKa = 11.84PIRR270 pKa = 11.84APRR273 pKa = 11.84PEE275 pKa = 4.24AMSITDD281 pKa = 3.43AAEE284 pKa = 3.83INVDD288 pKa = 3.43NFPNYY293 pKa = 8.71PANTGFNMQLLKK305 pKa = 10.82AVSSLIATTSTFKK318 pKa = 10.22VTSTNFSNLAEE329 pKa = 4.47TGSTAQAVLLQPIDD343 pKa = 3.83NDD345 pKa = 3.69DD346 pKa = 3.5PTIRR350 pKa = 11.84TINANLRR357 pKa = 11.84PQSLNHH363 pKa = 7.05DD364 pKa = 3.95PLATFGQAVAFGYY377 pKa = 10.42QLLKK381 pKa = 10.53EE382 pKa = 4.32PCGNNHH388 pKa = 6.43SAWCCITPAAGQVIPQQWIDD408 pKa = 3.38NRR410 pKa = 11.84NDD412 pKa = 3.0RR413 pKa = 11.84RR414 pKa = 11.84NIPAQFQVRR423 pKa = 11.84VFDD426 pKa = 4.43SNSVSAGDD434 pKa = 3.63YY435 pKa = 9.49RR436 pKa = 11.84RR437 pKa = 11.84KK438 pKa = 9.8IVEE441 pKa = 4.04KK442 pKa = 10.3LAKK445 pKa = 10.09KK446 pKa = 10.19SS447 pKa = 3.4
MM1 pKa = 8.17DD2 pKa = 5.04PRR4 pKa = 11.84PYY6 pKa = 10.33LGKK9 pKa = 9.35PANQKK14 pKa = 9.87NPRR17 pKa = 11.84AKK19 pKa = 10.08NLHH22 pKa = 5.37VPRR25 pKa = 11.84APRR28 pKa = 11.84EE29 pKa = 4.02APAAMEE35 pKa = 4.47DD36 pKa = 4.08TPPAPSKK43 pKa = 10.43QASKK47 pKa = 10.95PKK49 pKa = 9.76MPSTLPPPKK58 pKa = 9.72HH59 pKa = 6.48ASNPPRR65 pKa = 11.84KK66 pKa = 9.1EE67 pKa = 3.85DD68 pKa = 3.75FVEE71 pKa = 4.56KK72 pKa = 10.81SPLSDD77 pKa = 5.36AIFQEE82 pKa = 4.29DD83 pKa = 3.85MAVRR87 pKa = 11.84EE88 pKa = 4.37VNLTTRR94 pKa = 11.84QNVSLSAVNEE104 pKa = 4.09LSRR107 pKa = 11.84ATYY110 pKa = 9.94QQMLITDD117 pKa = 4.41PNLAKK122 pKa = 10.11QWTPEE127 pKa = 3.6VHH129 pKa = 7.18DD130 pKa = 4.5YY131 pKa = 11.36YY132 pKa = 10.12VTAMTWLRR140 pKa = 11.84IVALKK145 pKa = 10.63ASSGQDD151 pKa = 3.1LTPAEE156 pKa = 4.26EE157 pKa = 4.45TLLSMSATHH166 pKa = 6.49SFNLTEE172 pKa = 5.19PIRR175 pKa = 11.84LYY177 pKa = 11.09LSGVGVAVTKK187 pKa = 10.66NGQHH191 pKa = 7.28LYY193 pKa = 10.33PSFPPLPTEE202 pKa = 4.24VVDD205 pKa = 6.1GIPGLYY211 pKa = 10.38GPIGEE216 pKa = 4.56ATHH219 pKa = 6.32NLYY222 pKa = 10.9EE223 pKa = 4.73EE224 pKa = 4.55IPALGVSIGMIRR236 pKa = 11.84ASLNPVRR243 pKa = 11.84PIPQWVPPILPVNTNANPNLLGFRR267 pKa = 11.84PIRR270 pKa = 11.84APRR273 pKa = 11.84PEE275 pKa = 4.24AMSITDD281 pKa = 3.43AAEE284 pKa = 3.83INVDD288 pKa = 3.43NFPNYY293 pKa = 8.71PANTGFNMQLLKK305 pKa = 10.82AVSSLIATTSTFKK318 pKa = 10.22VTSTNFSNLAEE329 pKa = 4.47TGSTAQAVLLQPIDD343 pKa = 3.83NDD345 pKa = 3.69DD346 pKa = 3.5PTIRR350 pKa = 11.84TINANLRR357 pKa = 11.84PQSLNHH363 pKa = 7.05DD364 pKa = 3.95PLATFGQAVAFGYY377 pKa = 10.42QLLKK381 pKa = 10.53EE382 pKa = 4.32PCGNNHH388 pKa = 6.43SAWCCITPAAGQVIPQQWIDD408 pKa = 3.38NRR410 pKa = 11.84NDD412 pKa = 3.0RR413 pKa = 11.84RR414 pKa = 11.84NIPAQFQVRR423 pKa = 11.84VFDD426 pKa = 4.43SNSVSAGDD434 pKa = 3.63YY435 pKa = 9.49RR436 pKa = 11.84RR437 pKa = 11.84KK438 pKa = 9.8IVEE441 pKa = 4.04KK442 pKa = 10.3LAKK445 pKa = 10.09KK446 pKa = 10.19SS447 pKa = 3.4
Molecular weight: 49.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2649 |
338 |
543 |
441.5 |
49.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.72 ± 0.658 | 1.397 ± 0.19 |
5.436 ± 0.355 | 5.059 ± 0.12 |
3.473 ± 0.228 | 3.435 ± 0.431 |
3.662 ± 0.439 | 4.945 ± 0.359 |
4.379 ± 0.508 | 9.475 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.076 ± 0.156 | 4.341 ± 0.646 |
7.739 ± 0.704 | 4.19 ± 0.435 |
6.229 ± 0.458 | 7.324 ± 0.643 |
7.701 ± 0.707 | 5.7 ± 0.394 |
1.246 ± 0.231 | 3.473 ± 0.642 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
