
Circovirus-like genome RW-E
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
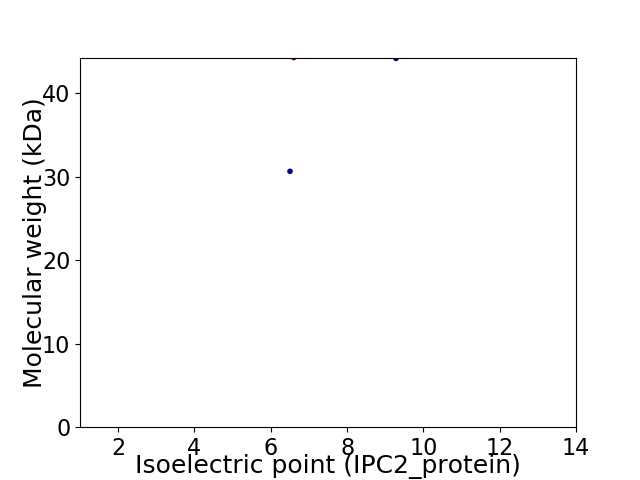
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GII4|C6GII4_9VIRU Uncharacterized protein OS=Circovirus-like genome RW-E OX=642255 PE=4 SV=1
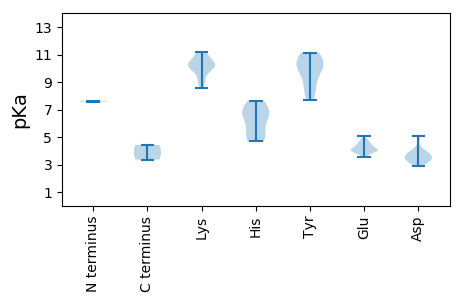
MM1 pKa = 7.56IRR3 pKa = 11.84RR4 pKa = 11.84QGVYY8 pKa = 8.65WLLTIPEE15 pKa = 4.73DD16 pKa = 3.56EE17 pKa = 5.05FEE19 pKa = 4.05PHH21 pKa = 6.91LPQDD25 pKa = 3.33CAYY28 pKa = 9.97IKK30 pKa = 9.99GQKK33 pKa = 8.96EE34 pKa = 4.04RR35 pKa = 11.84GEE37 pKa = 4.37HH38 pKa = 4.72TGYY41 pKa = 10.82LHH43 pKa = 5.62WQVLVVLRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 10.27GSLSTITGMFGVSCHH68 pKa = 6.66AEE70 pKa = 4.05LSRR73 pKa = 11.84SAAASEE79 pKa = 4.46YY80 pKa = 9.76VWKK83 pKa = 10.46EE84 pKa = 3.62DD85 pKa = 3.86TRR87 pKa = 11.84VGDD90 pKa = 3.42QFEE93 pKa = 4.67FGTLPFKK100 pKa = 11.15RR101 pKa = 11.84NDD103 pKa = 3.53PKK105 pKa = 10.98DD106 pKa = 3.19WEE108 pKa = 4.62QIWEE112 pKa = 4.02RR113 pKa = 11.84AKK115 pKa = 11.04AGDD118 pKa = 3.7INGIPADD125 pKa = 4.02VRR127 pKa = 11.84IQCYY131 pKa = 7.71RR132 pKa = 11.84TLRR135 pKa = 11.84TIRR138 pKa = 11.84ADD140 pKa = 3.57FAVPEE145 pKa = 4.08AMVRR149 pKa = 11.84QCFVYY154 pKa = 10.38CGPTGTGKK162 pKa = 10.01SRR164 pKa = 11.84RR165 pKa = 11.84AWAEE169 pKa = 3.52AGMDD173 pKa = 4.81AYY175 pKa = 10.13PKK177 pKa = 10.5DD178 pKa = 3.81PRR180 pKa = 11.84TKK182 pKa = 9.95FWDD185 pKa = 4.35GYY187 pKa = 10.16RR188 pKa = 11.84DD189 pKa = 3.57QKK191 pKa = 11.1HH192 pKa = 4.69VVVDD196 pKa = 4.05EE197 pKa = 4.0FRR199 pKa = 11.84GAIDD203 pKa = 3.08ISHH206 pKa = 7.45ILRR209 pKa = 11.84WLDD212 pKa = 3.42RR213 pKa = 11.84YY214 pKa = 7.9PTLVEE219 pKa = 3.92IKK221 pKa = 10.65GSATCLVAEE230 pKa = 4.76KK231 pKa = 10.52LWITSNVHH239 pKa = 5.12PNEE242 pKa = 4.31WYY244 pKa = 10.14PMLDD248 pKa = 3.25SATVDD253 pKa = 2.99ALLRR257 pKa = 11.84RR258 pKa = 11.84LQITVFEE265 pKa = 4.42
MM1 pKa = 7.56IRR3 pKa = 11.84RR4 pKa = 11.84QGVYY8 pKa = 8.65WLLTIPEE15 pKa = 4.73DD16 pKa = 3.56EE17 pKa = 5.05FEE19 pKa = 4.05PHH21 pKa = 6.91LPQDD25 pKa = 3.33CAYY28 pKa = 9.97IKK30 pKa = 9.99GQKK33 pKa = 8.96EE34 pKa = 4.04RR35 pKa = 11.84GEE37 pKa = 4.37HH38 pKa = 4.72TGYY41 pKa = 10.82LHH43 pKa = 5.62WQVLVVLRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 10.27GSLSTITGMFGVSCHH68 pKa = 6.66AEE70 pKa = 4.05LSRR73 pKa = 11.84SAAASEE79 pKa = 4.46YY80 pKa = 9.76VWKK83 pKa = 10.46EE84 pKa = 3.62DD85 pKa = 3.86TRR87 pKa = 11.84VGDD90 pKa = 3.42QFEE93 pKa = 4.67FGTLPFKK100 pKa = 11.15RR101 pKa = 11.84NDD103 pKa = 3.53PKK105 pKa = 10.98DD106 pKa = 3.19WEE108 pKa = 4.62QIWEE112 pKa = 4.02RR113 pKa = 11.84AKK115 pKa = 11.04AGDD118 pKa = 3.7INGIPADD125 pKa = 4.02VRR127 pKa = 11.84IQCYY131 pKa = 7.71RR132 pKa = 11.84TLRR135 pKa = 11.84TIRR138 pKa = 11.84ADD140 pKa = 3.57FAVPEE145 pKa = 4.08AMVRR149 pKa = 11.84QCFVYY154 pKa = 10.38CGPTGTGKK162 pKa = 10.01SRR164 pKa = 11.84RR165 pKa = 11.84AWAEE169 pKa = 3.52AGMDD173 pKa = 4.81AYY175 pKa = 10.13PKK177 pKa = 10.5DD178 pKa = 3.81PRR180 pKa = 11.84TKK182 pKa = 9.95FWDD185 pKa = 4.35GYY187 pKa = 10.16RR188 pKa = 11.84DD189 pKa = 3.57QKK191 pKa = 11.1HH192 pKa = 4.69VVVDD196 pKa = 4.05EE197 pKa = 4.0FRR199 pKa = 11.84GAIDD203 pKa = 3.08ISHH206 pKa = 7.45ILRR209 pKa = 11.84WLDD212 pKa = 3.42RR213 pKa = 11.84YY214 pKa = 7.9PTLVEE219 pKa = 3.92IKK221 pKa = 10.65GSATCLVAEE230 pKa = 4.76KK231 pKa = 10.52LWITSNVHH239 pKa = 5.12PNEE242 pKa = 4.31WYY244 pKa = 10.14PMLDD248 pKa = 3.25SATVDD253 pKa = 2.99ALLRR257 pKa = 11.84RR258 pKa = 11.84LQITVFEE265 pKa = 4.42
Molecular weight: 30.63 kDa
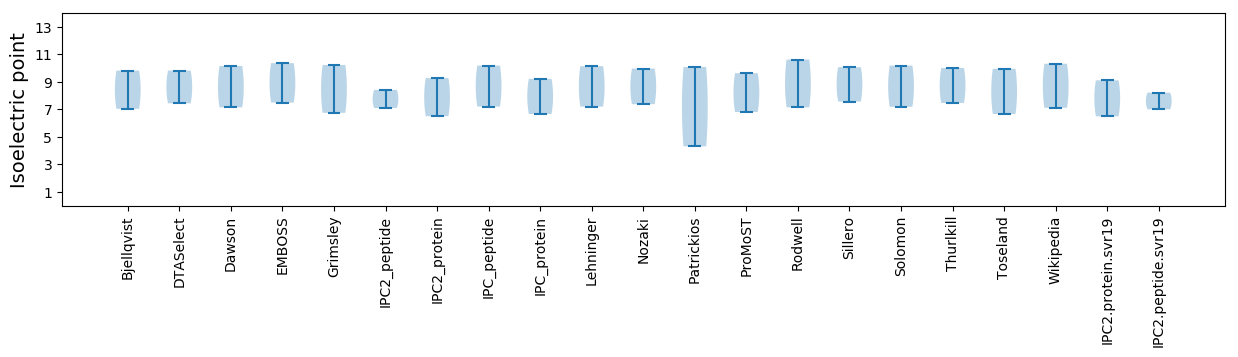
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GII4|C6GII4_9VIRU Uncharacterized protein OS=Circovirus-like genome RW-E OX=642255 PE=4 SV=1
MM1 pKa = 7.63PPIKK5 pKa = 9.84RR6 pKa = 11.84KK7 pKa = 10.32SPSTPKK13 pKa = 9.75SDD15 pKa = 2.86KK16 pKa = 10.15RR17 pKa = 11.84SRR19 pKa = 11.84GSITPGSARR28 pKa = 11.84RR29 pKa = 11.84VLFQARR35 pKa = 11.84VPRR38 pKa = 11.84QRR40 pKa = 11.84SGTNVVTGADD50 pKa = 3.2KK51 pKa = 11.09RR52 pKa = 11.84IGKK55 pKa = 9.07KK56 pKa = 8.75VKK58 pKa = 10.19KK59 pKa = 8.96EE60 pKa = 3.85GRR62 pKa = 11.84VKK64 pKa = 10.52RR65 pKa = 11.84LKK67 pKa = 10.11VSKK70 pKa = 10.64AFRR73 pKa = 11.84KK74 pKa = 9.43KK75 pKa = 10.2VKK77 pKa = 10.21QSLEE81 pKa = 3.98THH83 pKa = 6.96RR84 pKa = 11.84GTGWFRR90 pKa = 11.84EE91 pKa = 4.21TVTIEE96 pKa = 3.97KK97 pKa = 10.16LNCIDD102 pKa = 3.47SQQNVLTPGRR112 pKa = 11.84QVNGTFGNFFSPTYY126 pKa = 8.84IRR128 pKa = 11.84YY129 pKa = 8.79VASHH133 pKa = 7.46LYY135 pKa = 10.43NKK137 pKa = 9.56LAPTQAITVNDD148 pKa = 3.8TNLYY152 pKa = 9.36PVANLKK158 pKa = 10.03IDD160 pKa = 4.09VIEE163 pKa = 3.94QNYY166 pKa = 8.58VMKK169 pKa = 10.69LRR171 pKa = 11.84NNSPRR176 pKa = 11.84TYY178 pKa = 10.99DD179 pKa = 3.35VTLVDD184 pKa = 5.07YY185 pKa = 10.91SPKK188 pKa = 10.31SNNSIATSFEE198 pKa = 4.27DD199 pKa = 3.17TWTSALVNGGPFGAAGTLGQEE220 pKa = 3.94SRR222 pKa = 11.84EE223 pKa = 4.07NVGNATKK230 pKa = 9.57NTMGNHH236 pKa = 6.1PKK238 pKa = 10.14FMAVMRR244 pKa = 11.84HH245 pKa = 5.36YY246 pKa = 11.12YY247 pKa = 11.13SMDD250 pKa = 3.27TVHH253 pKa = 6.75VKK255 pKa = 10.76LEE257 pKa = 4.17PGKK260 pKa = 10.52EE261 pKa = 4.08YY262 pKa = 9.88YY263 pKa = 10.49HH264 pKa = 6.47KK265 pKa = 11.15VKK267 pKa = 9.99GPNMMTYY274 pKa = 11.04DD275 pKa = 3.49FNKK278 pKa = 10.06YY279 pKa = 9.81FKK281 pKa = 10.51DD282 pKa = 3.89SNFHH286 pKa = 7.63DD287 pKa = 3.25IQKK290 pKa = 9.63FCKK293 pKa = 8.58GTLVMCSLDD302 pKa = 3.68LTSTTLATHH311 pKa = 6.49GRR313 pKa = 11.84YY314 pKa = 7.93TDD316 pKa = 3.79IPTADD321 pKa = 3.83PQGMIIEE328 pKa = 4.59TTHH331 pKa = 4.87FTKK334 pKa = 10.49IKK336 pKa = 9.21CPEE339 pKa = 3.78RR340 pKa = 11.84AGFQIPNVAVPIGSIQPLSQQSQNAWSIVYY370 pKa = 9.65PGTAQAGVVEE380 pKa = 4.3YY381 pKa = 10.62RR382 pKa = 11.84QDD384 pKa = 3.71EE385 pKa = 4.48NPLSNVIEE393 pKa = 4.29VV394 pKa = 3.36
MM1 pKa = 7.63PPIKK5 pKa = 9.84RR6 pKa = 11.84KK7 pKa = 10.32SPSTPKK13 pKa = 9.75SDD15 pKa = 2.86KK16 pKa = 10.15RR17 pKa = 11.84SRR19 pKa = 11.84GSITPGSARR28 pKa = 11.84RR29 pKa = 11.84VLFQARR35 pKa = 11.84VPRR38 pKa = 11.84QRR40 pKa = 11.84SGTNVVTGADD50 pKa = 3.2KK51 pKa = 11.09RR52 pKa = 11.84IGKK55 pKa = 9.07KK56 pKa = 8.75VKK58 pKa = 10.19KK59 pKa = 8.96EE60 pKa = 3.85GRR62 pKa = 11.84VKK64 pKa = 10.52RR65 pKa = 11.84LKK67 pKa = 10.11VSKK70 pKa = 10.64AFRR73 pKa = 11.84KK74 pKa = 9.43KK75 pKa = 10.2VKK77 pKa = 10.21QSLEE81 pKa = 3.98THH83 pKa = 6.96RR84 pKa = 11.84GTGWFRR90 pKa = 11.84EE91 pKa = 4.21TVTIEE96 pKa = 3.97KK97 pKa = 10.16LNCIDD102 pKa = 3.47SQQNVLTPGRR112 pKa = 11.84QVNGTFGNFFSPTYY126 pKa = 8.84IRR128 pKa = 11.84YY129 pKa = 8.79VASHH133 pKa = 7.46LYY135 pKa = 10.43NKK137 pKa = 9.56LAPTQAITVNDD148 pKa = 3.8TNLYY152 pKa = 9.36PVANLKK158 pKa = 10.03IDD160 pKa = 4.09VIEE163 pKa = 3.94QNYY166 pKa = 8.58VMKK169 pKa = 10.69LRR171 pKa = 11.84NNSPRR176 pKa = 11.84TYY178 pKa = 10.99DD179 pKa = 3.35VTLVDD184 pKa = 5.07YY185 pKa = 10.91SPKK188 pKa = 10.31SNNSIATSFEE198 pKa = 4.27DD199 pKa = 3.17TWTSALVNGGPFGAAGTLGQEE220 pKa = 3.94SRR222 pKa = 11.84EE223 pKa = 4.07NVGNATKK230 pKa = 9.57NTMGNHH236 pKa = 6.1PKK238 pKa = 10.14FMAVMRR244 pKa = 11.84HH245 pKa = 5.36YY246 pKa = 11.12YY247 pKa = 11.13SMDD250 pKa = 3.27TVHH253 pKa = 6.75VKK255 pKa = 10.76LEE257 pKa = 4.17PGKK260 pKa = 10.52EE261 pKa = 4.08YY262 pKa = 9.88YY263 pKa = 10.49HH264 pKa = 6.47KK265 pKa = 11.15VKK267 pKa = 9.99GPNMMTYY274 pKa = 11.04DD275 pKa = 3.49FNKK278 pKa = 10.06YY279 pKa = 9.81FKK281 pKa = 10.51DD282 pKa = 3.89SNFHH286 pKa = 7.63DD287 pKa = 3.25IQKK290 pKa = 9.63FCKK293 pKa = 8.58GTLVMCSLDD302 pKa = 3.68LTSTTLATHH311 pKa = 6.49GRR313 pKa = 11.84YY314 pKa = 7.93TDD316 pKa = 3.79IPTADD321 pKa = 3.83PQGMIIEE328 pKa = 4.59TTHH331 pKa = 4.87FTKK334 pKa = 10.49IKK336 pKa = 9.21CPEE339 pKa = 3.78RR340 pKa = 11.84AGFQIPNVAVPIGSIQPLSQQSQNAWSIVYY370 pKa = 9.65PGTAQAGVVEE380 pKa = 4.3YY381 pKa = 10.62RR382 pKa = 11.84QDD384 pKa = 3.71EE385 pKa = 4.48NPLSNVIEE393 pKa = 4.29VV394 pKa = 3.36
Molecular weight: 44.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
659 |
265 |
394 |
329.5 |
37.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.222 ± 0.788 | 1.517 ± 0.444 |
5.159 ± 0.97 | 5.008 ± 1.06 |
3.794 ± 0.012 | 6.829 ± 0.021 |
2.428 ± 0.127 | 5.311 ± 0.208 |
6.829 ± 1.142 | 5.918 ± 0.744 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.231 | 4.552 ± 1.808 |
5.615 ± 0.421 | 4.249 ± 0.282 |
6.98 ± 1.009 | 5.918 ± 1.05 |
7.739 ± 1.011 | 7.739 ± 0.338 |
1.973 ± 1.07 | 3.945 ± 0.102 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
