
Bombyx mori densovirus 5 (isolate Shinshu) (BmDNV-5)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Densovirinae; Iteradensovirus; Lepidopteran iteradensovirus 1; Bombyx mori densovirus; Bombyx mori densovirus 5
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
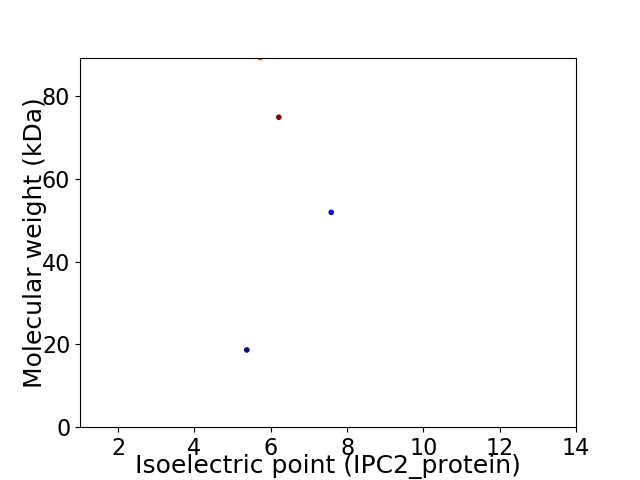
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
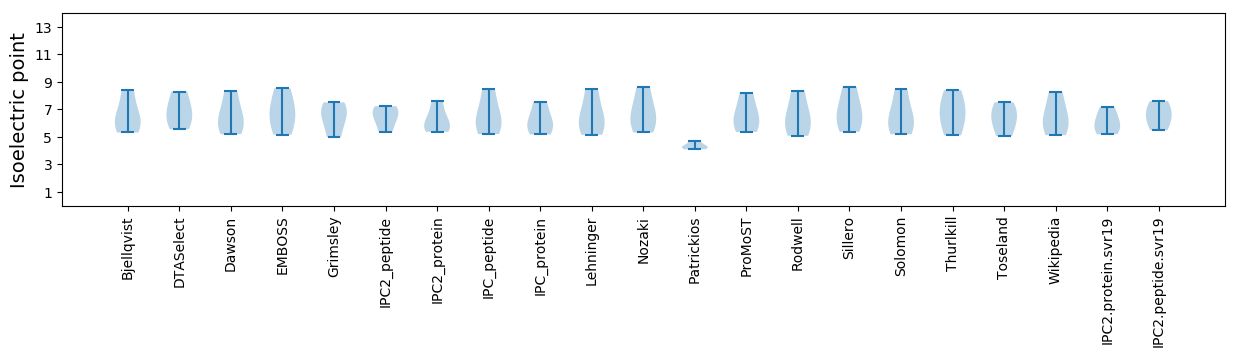
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9JFY3|Q9JFY3_BMDNS Nonstructural protein OS=Bombyx mori densovirus 5 (isolate Shinshu) OX=648251 GN=orf1 PE=4 SV=1
MM1 pKa = 7.08FRR3 pKa = 11.84VDD5 pKa = 5.24RR6 pKa = 11.84VDD8 pKa = 3.85RR9 pKa = 11.84DD10 pKa = 3.97CLILPWIDD18 pKa = 3.14VKK20 pKa = 11.27APPEE24 pKa = 4.15APAFIIVPSAGVKK37 pKa = 8.89YY38 pKa = 10.89VEE40 pKa = 4.59MALGWFFKK48 pKa = 11.02LFFFVSTPFDD58 pKa = 3.57VVIDD62 pKa = 3.58FLFRR66 pKa = 11.84VVFICLEE73 pKa = 3.79LDD75 pKa = 3.37GRR77 pKa = 11.84FLRR80 pKa = 11.84FSSSLSDD87 pKa = 5.29SDD89 pKa = 5.75WLDD92 pKa = 3.16CWLSAVSVSSRR103 pKa = 11.84YY104 pKa = 9.95DD105 pKa = 3.24SEE107 pKa = 5.18IVVGWVSSTNVFWVVFRR124 pKa = 11.84SFICFMEE131 pKa = 4.7PNSVRR136 pKa = 11.84TSAIKK141 pKa = 10.45SSPDD145 pKa = 3.33FVNHH149 pKa = 5.4FTKK152 pKa = 10.65SVAWSKK158 pKa = 11.47LNGVVV163 pKa = 3.63
MM1 pKa = 7.08FRR3 pKa = 11.84VDD5 pKa = 5.24RR6 pKa = 11.84VDD8 pKa = 3.85RR9 pKa = 11.84DD10 pKa = 3.97CLILPWIDD18 pKa = 3.14VKK20 pKa = 11.27APPEE24 pKa = 4.15APAFIIVPSAGVKK37 pKa = 8.89YY38 pKa = 10.89VEE40 pKa = 4.59MALGWFFKK48 pKa = 11.02LFFFVSTPFDD58 pKa = 3.57VVIDD62 pKa = 3.58FLFRR66 pKa = 11.84VVFICLEE73 pKa = 3.79LDD75 pKa = 3.37GRR77 pKa = 11.84FLRR80 pKa = 11.84FSSSLSDD87 pKa = 5.29SDD89 pKa = 5.75WLDD92 pKa = 3.16CWLSAVSVSSRR103 pKa = 11.84YY104 pKa = 9.95DD105 pKa = 3.24SEE107 pKa = 5.18IVVGWVSSTNVFWVVFRR124 pKa = 11.84SFICFMEE131 pKa = 4.7PNSVRR136 pKa = 11.84TSAIKK141 pKa = 10.45SSPDD145 pKa = 3.33FVNHH149 pKa = 5.4FTKK152 pKa = 10.65SVAWSKK158 pKa = 11.47LNGVVV163 pKa = 3.63
Molecular weight: 18.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9JFY0|Q9JFY0_BMDNS Capsid protein OS=Bombyx mori densovirus 5 (isolate Shinshu) OX=648251 GN=orf2 PE=4 SV=1
MM1 pKa = 7.84LILLVNFCLLWSLLKK16 pKa = 10.46SLQMKK21 pKa = 8.41FQVKK25 pKa = 8.36QRR27 pKa = 11.84PEE29 pKa = 3.83YY30 pKa = 9.32LQKK33 pKa = 11.04LEE35 pKa = 4.17TVMMSTIQDD44 pKa = 3.6WDD46 pKa = 3.34TMMKK50 pKa = 9.93VVSAEE55 pKa = 3.79EE56 pKa = 3.76EE57 pKa = 4.04MIEE60 pKa = 4.14FLTEE64 pKa = 3.62EE65 pKa = 4.61FSAFIQKK72 pKa = 9.81DD73 pKa = 3.73CKK75 pKa = 10.93NNSLEE80 pKa = 4.08EE81 pKa = 4.24LYY83 pKa = 9.59EE84 pKa = 4.27TVDD87 pKa = 3.57EE88 pKa = 4.72PKK90 pKa = 10.5LVHH93 pKa = 7.04SSLATMKK100 pKa = 10.82YY101 pKa = 9.46YY102 pKa = 9.86LTLILQNKK110 pKa = 8.8AKK112 pKa = 8.6EE113 pKa = 4.28TTDD116 pKa = 3.59LKK118 pKa = 10.97GHH120 pKa = 5.75SLSVINKK127 pKa = 6.79WNSVIKK133 pKa = 9.91TPLKK137 pKa = 10.34LLSNEE142 pKa = 4.48KK143 pKa = 9.85EE144 pKa = 3.72ASTNSFEE151 pKa = 4.08RR152 pKa = 11.84WNAEE156 pKa = 3.78WEE158 pKa = 4.34SSQISYY164 pKa = 11.4AEE166 pKa = 4.19LLRR169 pKa = 11.84IIQLMQEE176 pKa = 4.18EE177 pKa = 6.19FYY179 pKa = 8.18TTPLSLDD186 pKa = 3.17QATDD190 pKa = 3.63FVKK193 pKa = 10.18WFTKK197 pKa = 10.39SGEE200 pKa = 3.94LLIAEE205 pKa = 4.31VRR207 pKa = 11.84TLFGSMKK214 pKa = 9.93QIKK217 pKa = 8.26EE218 pKa = 3.87RR219 pKa = 11.84KK220 pKa = 4.88TTQKK224 pKa = 9.55TLVEE228 pKa = 4.37DD229 pKa = 3.7THH231 pKa = 6.54PTTISEE237 pKa = 4.47SYY239 pKa = 10.89RR240 pKa = 11.84LDD242 pKa = 3.23TDD244 pKa = 3.67TAEE247 pKa = 4.59SQQSSQSEE255 pKa = 4.37SEE257 pKa = 4.01RR258 pKa = 11.84EE259 pKa = 3.73EE260 pKa = 3.79EE261 pKa = 4.0NRR263 pKa = 11.84RR264 pKa = 11.84NRR266 pKa = 11.84PSSSRR271 pKa = 11.84HH272 pKa = 4.68INTTRR277 pKa = 11.84KK278 pKa = 9.46RR279 pKa = 11.84KK280 pKa = 10.31SITTSKK286 pKa = 11.08GVLTKK291 pKa = 10.56KK292 pKa = 10.4KK293 pKa = 10.21SLKK296 pKa = 8.88NQPRR300 pKa = 11.84AISTYY305 pKa = 7.54FTPADD310 pKa = 3.77GTIMNAGASGGALTSIQGRR329 pKa = 11.84IRR331 pKa = 11.84QSLSTRR337 pKa = 11.84STRR340 pKa = 11.84NIYY343 pKa = 8.82ATLCSITQNGPDD355 pKa = 4.12GLCTSKK361 pKa = 10.41WPTDD365 pKa = 3.64PNSNMFIEE373 pKa = 4.83LNLFAKK379 pKa = 9.9NQYY382 pKa = 9.77NPSDD386 pKa = 3.35TRR388 pKa = 11.84NNWKK392 pKa = 10.11RR393 pKa = 11.84ATIRR397 pKa = 11.84MKK399 pKa = 10.76VDD401 pKa = 3.24LPDD404 pKa = 3.83NSRR407 pKa = 11.84HH408 pKa = 5.81AYY410 pKa = 7.51QLKK413 pKa = 10.32EE414 pKa = 4.09IIDD417 pKa = 3.91AQMSRR422 pKa = 11.84MEE424 pKa = 4.46DD425 pKa = 3.26HH426 pKa = 7.62PEE428 pKa = 3.72TEE430 pKa = 4.56TQSKK434 pKa = 10.77SSWSNSNSGQHH445 pKa = 6.23GPWNTYY451 pKa = 8.41
MM1 pKa = 7.84LILLVNFCLLWSLLKK16 pKa = 10.46SLQMKK21 pKa = 8.41FQVKK25 pKa = 8.36QRR27 pKa = 11.84PEE29 pKa = 3.83YY30 pKa = 9.32LQKK33 pKa = 11.04LEE35 pKa = 4.17TVMMSTIQDD44 pKa = 3.6WDD46 pKa = 3.34TMMKK50 pKa = 9.93VVSAEE55 pKa = 3.79EE56 pKa = 3.76EE57 pKa = 4.04MIEE60 pKa = 4.14FLTEE64 pKa = 3.62EE65 pKa = 4.61FSAFIQKK72 pKa = 9.81DD73 pKa = 3.73CKK75 pKa = 10.93NNSLEE80 pKa = 4.08EE81 pKa = 4.24LYY83 pKa = 9.59EE84 pKa = 4.27TVDD87 pKa = 3.57EE88 pKa = 4.72PKK90 pKa = 10.5LVHH93 pKa = 7.04SSLATMKK100 pKa = 10.82YY101 pKa = 9.46YY102 pKa = 9.86LTLILQNKK110 pKa = 8.8AKK112 pKa = 8.6EE113 pKa = 4.28TTDD116 pKa = 3.59LKK118 pKa = 10.97GHH120 pKa = 5.75SLSVINKK127 pKa = 6.79WNSVIKK133 pKa = 9.91TPLKK137 pKa = 10.34LLSNEE142 pKa = 4.48KK143 pKa = 9.85EE144 pKa = 3.72ASTNSFEE151 pKa = 4.08RR152 pKa = 11.84WNAEE156 pKa = 3.78WEE158 pKa = 4.34SSQISYY164 pKa = 11.4AEE166 pKa = 4.19LLRR169 pKa = 11.84IIQLMQEE176 pKa = 4.18EE177 pKa = 6.19FYY179 pKa = 8.18TTPLSLDD186 pKa = 3.17QATDD190 pKa = 3.63FVKK193 pKa = 10.18WFTKK197 pKa = 10.39SGEE200 pKa = 3.94LLIAEE205 pKa = 4.31VRR207 pKa = 11.84TLFGSMKK214 pKa = 9.93QIKK217 pKa = 8.26EE218 pKa = 3.87RR219 pKa = 11.84KK220 pKa = 4.88TTQKK224 pKa = 9.55TLVEE228 pKa = 4.37DD229 pKa = 3.7THH231 pKa = 6.54PTTISEE237 pKa = 4.47SYY239 pKa = 10.89RR240 pKa = 11.84LDD242 pKa = 3.23TDD244 pKa = 3.67TAEE247 pKa = 4.59SQQSSQSEE255 pKa = 4.37SEE257 pKa = 4.01RR258 pKa = 11.84EE259 pKa = 3.73EE260 pKa = 3.79EE261 pKa = 4.0NRR263 pKa = 11.84RR264 pKa = 11.84NRR266 pKa = 11.84PSSSRR271 pKa = 11.84HH272 pKa = 4.68INTTRR277 pKa = 11.84KK278 pKa = 9.46RR279 pKa = 11.84KK280 pKa = 10.31SITTSKK286 pKa = 11.08GVLTKK291 pKa = 10.56KK292 pKa = 10.4KK293 pKa = 10.21SLKK296 pKa = 8.88NQPRR300 pKa = 11.84AISTYY305 pKa = 7.54FTPADD310 pKa = 3.77GTIMNAGASGGALTSIQGRR329 pKa = 11.84IRR331 pKa = 11.84QSLSTRR337 pKa = 11.84STRR340 pKa = 11.84NIYY343 pKa = 8.82ATLCSITQNGPDD355 pKa = 4.12GLCTSKK361 pKa = 10.41WPTDD365 pKa = 3.64PNSNMFIEE373 pKa = 4.83LNLFAKK379 pKa = 9.9NQYY382 pKa = 9.77NPSDD386 pKa = 3.35TRR388 pKa = 11.84NNWKK392 pKa = 10.11RR393 pKa = 11.84ATIRR397 pKa = 11.84MKK399 pKa = 10.76VDD401 pKa = 3.24LPDD404 pKa = 3.83NSRR407 pKa = 11.84HH408 pKa = 5.81AYY410 pKa = 7.51QLKK413 pKa = 10.32EE414 pKa = 4.09IIDD417 pKa = 3.91AQMSRR422 pKa = 11.84MEE424 pKa = 4.46DD425 pKa = 3.26HH426 pKa = 7.62PEE428 pKa = 3.72TEE430 pKa = 4.56TQSKK434 pKa = 10.77SSWSNSNSGQHH445 pKa = 6.23GPWNTYY451 pKa = 8.41
Molecular weight: 51.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2040 |
163 |
754 |
510.0 |
58.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.539 ± 0.862 | 1.127 ± 0.32 |
5.539 ± 0.528 | 7.402 ± 1.202 |
4.853 ± 1.312 | 5.098 ± 1.083 |
2.157 ± 0.365 | 5.637 ± 0.149 |
5.98 ± 0.64 | 7.108 ± 0.693 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.157 ± 0.329 | 5.931 ± 0.691 |
5.147 ± 0.564 | 4.216 ± 0.823 |
6.275 ± 0.67 | 7.5 ± 1.588 |
6.863 ± 1.18 | 5.49 ± 1.761 |
1.863 ± 0.518 | 4.118 ± 0.837 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
