
Aquimarina spongiae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
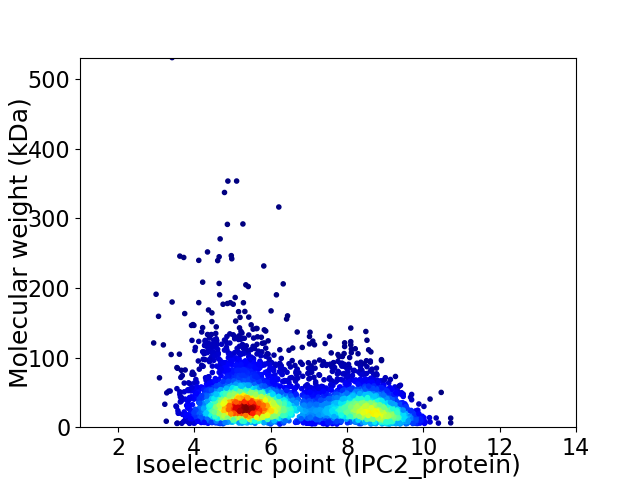
Virtual 2D-PAGE plot for 4714 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6A1S5|A0A1M6A1S5_9FLAO Arylsulfatase A OS=Aquimarina spongiae OX=570521 GN=SAMN04488508_1015 PE=3 SV=1
PP1 pKa = 6.97MPDD4 pKa = 2.7ITVVTDD10 pKa = 3.32EE11 pKa = 5.29ADD13 pKa = 3.51NCSSSPVVAFVEE25 pKa = 5.12DD26 pKa = 3.64ISDD29 pKa = 3.75GNSNPEE35 pKa = 4.22VITRR39 pKa = 11.84IYY41 pKa = 10.83SVTDD45 pKa = 3.23AAGNSINVSQTITINDD61 pKa = 3.67TTFPTASNPSSISAQCSAPMPDD83 pKa = 2.66ITVVTDD89 pKa = 3.31EE90 pKa = 5.43ADD92 pKa = 3.37NCSTPNVAFVSDD104 pKa = 4.72VITSPGIITRR114 pKa = 11.84TYY116 pKa = 10.43SVTDD120 pKa = 3.38AAEE123 pKa = 3.63NSINVTQTITLNDD136 pKa = 4.14NILPTASNLPPINAQCSAPAPDD158 pKa = 3.19ITIITDD164 pKa = 3.17HH165 pKa = 7.11TDD167 pKa = 2.88NCGTPTIAFVNDD179 pKa = 3.25VSDD182 pKa = 4.18GNSNPEE188 pKa = 4.24VITRR192 pKa = 11.84TYY194 pKa = 11.12SVTDD198 pKa = 3.4AAGNSINVTQIITINDD214 pKa = 3.62GTDD217 pKa = 3.28PNINCPANITQNADD231 pKa = 3.28ANSTFATVVYY241 pKa = 10.34SDD243 pKa = 5.23PIVNDD248 pKa = 3.17NCGVNSIVQLTGLPSGAEE266 pKa = 3.97FPIGTTINTFVLTDD280 pKa = 3.47NAGNTSQCSFEE291 pKa = 4.18VTVIDD296 pKa = 5.44NPLPTCTIDD305 pKa = 3.06AGEE308 pKa = 4.6DD309 pKa = 3.39EE310 pKa = 5.7RR311 pKa = 11.84ITEE314 pKa = 4.12GEE316 pKa = 4.35EE317 pKa = 3.66IQLDD321 pKa = 3.83ATASTTGSFVWSPSIGLSDD340 pKa = 3.58TTTSNPIASPSITTTYY356 pKa = 10.48LVEE359 pKa = 4.28FTSEE363 pKa = 4.12DD364 pKa = 3.36GCVAVDD370 pKa = 3.47EE371 pKa = 5.14VIVFVTPQEE380 pKa = 4.08EE381 pKa = 4.35DD382 pKa = 2.87EE383 pKa = 4.32TRR385 pKa = 11.84YY386 pKa = 10.68GFSPDD391 pKa = 2.99GDD393 pKa = 4.16GINEE397 pKa = 3.9FWEE400 pKa = 3.89IDD402 pKa = 3.7TIEE405 pKa = 4.46NYY407 pKa = 10.0PNNSVSIFNRR417 pKa = 11.84WGDD420 pKa = 3.53LVFEE424 pKa = 4.26IEE426 pKa = 4.78GYY428 pKa = 10.9NNTSRR433 pKa = 11.84VFRR436 pKa = 11.84GIANRR441 pKa = 11.84KK442 pKa = 8.65RR443 pKa = 11.84SLGGDD448 pKa = 3.34QLPEE452 pKa = 3.48GTYY455 pKa = 9.89FFKK458 pKa = 10.67IRR460 pKa = 11.84TSGPNSLRR468 pKa = 11.84KK469 pKa = 8.35TEE471 pKa = 4.14GFLVLKK477 pKa = 10.6RR478 pKa = 3.84
PP1 pKa = 6.97MPDD4 pKa = 2.7ITVVTDD10 pKa = 3.32EE11 pKa = 5.29ADD13 pKa = 3.51NCSSSPVVAFVEE25 pKa = 5.12DD26 pKa = 3.64ISDD29 pKa = 3.75GNSNPEE35 pKa = 4.22VITRR39 pKa = 11.84IYY41 pKa = 10.83SVTDD45 pKa = 3.23AAGNSINVSQTITINDD61 pKa = 3.67TTFPTASNPSSISAQCSAPMPDD83 pKa = 2.66ITVVTDD89 pKa = 3.31EE90 pKa = 5.43ADD92 pKa = 3.37NCSTPNVAFVSDD104 pKa = 4.72VITSPGIITRR114 pKa = 11.84TYY116 pKa = 10.43SVTDD120 pKa = 3.38AAEE123 pKa = 3.63NSINVTQTITLNDD136 pKa = 4.14NILPTASNLPPINAQCSAPAPDD158 pKa = 3.19ITIITDD164 pKa = 3.17HH165 pKa = 7.11TDD167 pKa = 2.88NCGTPTIAFVNDD179 pKa = 3.25VSDD182 pKa = 4.18GNSNPEE188 pKa = 4.24VITRR192 pKa = 11.84TYY194 pKa = 11.12SVTDD198 pKa = 3.4AAGNSINVTQIITINDD214 pKa = 3.62GTDD217 pKa = 3.28PNINCPANITQNADD231 pKa = 3.28ANSTFATVVYY241 pKa = 10.34SDD243 pKa = 5.23PIVNDD248 pKa = 3.17NCGVNSIVQLTGLPSGAEE266 pKa = 3.97FPIGTTINTFVLTDD280 pKa = 3.47NAGNTSQCSFEE291 pKa = 4.18VTVIDD296 pKa = 5.44NPLPTCTIDD305 pKa = 3.06AGEE308 pKa = 4.6DD309 pKa = 3.39EE310 pKa = 5.7RR311 pKa = 11.84ITEE314 pKa = 4.12GEE316 pKa = 4.35EE317 pKa = 3.66IQLDD321 pKa = 3.83ATASTTGSFVWSPSIGLSDD340 pKa = 3.58TTTSNPIASPSITTTYY356 pKa = 10.48LVEE359 pKa = 4.28FTSEE363 pKa = 4.12DD364 pKa = 3.36GCVAVDD370 pKa = 3.47EE371 pKa = 5.14VIVFVTPQEE380 pKa = 4.08EE381 pKa = 4.35DD382 pKa = 2.87EE383 pKa = 4.32TRR385 pKa = 11.84YY386 pKa = 10.68GFSPDD391 pKa = 2.99GDD393 pKa = 4.16GINEE397 pKa = 3.9FWEE400 pKa = 3.89IDD402 pKa = 3.7TIEE405 pKa = 4.46NYY407 pKa = 10.0PNNSVSIFNRR417 pKa = 11.84WGDD420 pKa = 3.53LVFEE424 pKa = 4.26IEE426 pKa = 4.78GYY428 pKa = 10.9NNTSRR433 pKa = 11.84VFRR436 pKa = 11.84GIANRR441 pKa = 11.84KK442 pKa = 8.65RR443 pKa = 11.84SLGGDD448 pKa = 3.34QLPEE452 pKa = 3.48GTYY455 pKa = 9.89FFKK458 pKa = 10.67IRR460 pKa = 11.84TSGPNSLRR468 pKa = 11.84KK469 pKa = 8.35TEE471 pKa = 4.14GFLVLKK477 pKa = 10.6RR478 pKa = 3.84
Molecular weight: 50.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6EC64|A0A1M6EC64_9FLAO Elongation factor Ts OS=Aquimarina spongiae OX=570521 GN=tsf PE=3 SV=1
MM1 pKa = 7.51KK2 pKa = 10.59SKK4 pKa = 9.01ITLILIICLATTHH17 pKa = 7.14LFAQEE22 pKa = 4.54DD23 pKa = 3.94LKK25 pKa = 11.52LWTGYY30 pKa = 10.23SAGARR35 pKa = 11.84ITKK38 pKa = 7.99EE39 pKa = 3.57WRR41 pKa = 11.84VSAGQLLLFSQQPFEE56 pKa = 4.29LSSMQNSVNLTYY68 pKa = 10.76RR69 pKa = 11.84FDD71 pKa = 3.45RR72 pKa = 11.84RR73 pKa = 11.84FRR75 pKa = 11.84AGFGYY80 pKa = 10.35LRR82 pKa = 11.84SSDD85 pKa = 3.93PSDD88 pKa = 3.95PDD90 pKa = 3.0QEE92 pKa = 3.98ARR94 pKa = 11.84NRR96 pKa = 11.84VTGRR100 pKa = 11.84FRR102 pKa = 11.84YY103 pKa = 9.1NARR106 pKa = 11.84LGKK109 pKa = 10.0FRR111 pKa = 11.84FLNSLRR117 pKa = 11.84AEE119 pKa = 3.69WHH121 pKa = 5.51FPEE124 pKa = 4.94RR125 pKa = 11.84SKK127 pKa = 11.26FEE129 pKa = 3.41YY130 pKa = 10.06RR131 pKa = 11.84IRR133 pKa = 11.84YY134 pKa = 7.84GIRR137 pKa = 11.84IHH139 pKa = 6.49RR140 pKa = 11.84GSWGLPLRR148 pKa = 11.84TTPFITNEE156 pKa = 3.27FHH158 pKa = 7.04YY159 pKa = 9.69YY160 pKa = 10.4LSGRR164 pKa = 11.84PLQYY168 pKa = 9.87RR169 pKa = 11.84DD170 pKa = 3.44EE171 pKa = 4.3QGEE174 pKa = 4.45KK175 pKa = 10.33VVKK178 pKa = 10.27QSPDD182 pKa = 2.78GLHH185 pKa = 6.03AHH187 pKa = 7.73RR188 pKa = 11.84ITLGVRR194 pKa = 11.84FRR196 pKa = 11.84PFKK199 pKa = 10.36RR200 pKa = 11.84ANASISYY207 pKa = 8.32MRR209 pKa = 11.84QTEE212 pKa = 3.93FNIGSKK218 pKa = 9.99YY219 pKa = 10.35RR220 pKa = 11.84EE221 pKa = 4.16INVTDD226 pKa = 4.14PRR228 pKa = 11.84DD229 pKa = 3.56GDD231 pKa = 3.53ILRR234 pKa = 11.84AFNNFSVLMFRR245 pKa = 11.84FSYY248 pKa = 10.67RR249 pKa = 11.84LDD251 pKa = 3.23FRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84TLRR258 pKa = 11.84PNAIGFHH265 pKa = 5.38TKK267 pKa = 8.6KK268 pKa = 10.28QKK270 pKa = 10.25PIPHH274 pKa = 6.2SEE276 pKa = 4.1EE277 pKa = 3.9SHH279 pKa = 5.65TSII282 pKa = 5.37
MM1 pKa = 7.51KK2 pKa = 10.59SKK4 pKa = 9.01ITLILIICLATTHH17 pKa = 7.14LFAQEE22 pKa = 4.54DD23 pKa = 3.94LKK25 pKa = 11.52LWTGYY30 pKa = 10.23SAGARR35 pKa = 11.84ITKK38 pKa = 7.99EE39 pKa = 3.57WRR41 pKa = 11.84VSAGQLLLFSQQPFEE56 pKa = 4.29LSSMQNSVNLTYY68 pKa = 10.76RR69 pKa = 11.84FDD71 pKa = 3.45RR72 pKa = 11.84RR73 pKa = 11.84FRR75 pKa = 11.84AGFGYY80 pKa = 10.35LRR82 pKa = 11.84SSDD85 pKa = 3.93PSDD88 pKa = 3.95PDD90 pKa = 3.0QEE92 pKa = 3.98ARR94 pKa = 11.84NRR96 pKa = 11.84VTGRR100 pKa = 11.84FRR102 pKa = 11.84YY103 pKa = 9.1NARR106 pKa = 11.84LGKK109 pKa = 10.0FRR111 pKa = 11.84FLNSLRR117 pKa = 11.84AEE119 pKa = 3.69WHH121 pKa = 5.51FPEE124 pKa = 4.94RR125 pKa = 11.84SKK127 pKa = 11.26FEE129 pKa = 3.41YY130 pKa = 10.06RR131 pKa = 11.84IRR133 pKa = 11.84YY134 pKa = 7.84GIRR137 pKa = 11.84IHH139 pKa = 6.49RR140 pKa = 11.84GSWGLPLRR148 pKa = 11.84TTPFITNEE156 pKa = 3.27FHH158 pKa = 7.04YY159 pKa = 9.69YY160 pKa = 10.4LSGRR164 pKa = 11.84PLQYY168 pKa = 9.87RR169 pKa = 11.84DD170 pKa = 3.44EE171 pKa = 4.3QGEE174 pKa = 4.45KK175 pKa = 10.33VVKK178 pKa = 10.27QSPDD182 pKa = 2.78GLHH185 pKa = 6.03AHH187 pKa = 7.73RR188 pKa = 11.84ITLGVRR194 pKa = 11.84FRR196 pKa = 11.84PFKK199 pKa = 10.36RR200 pKa = 11.84ANASISYY207 pKa = 8.32MRR209 pKa = 11.84QTEE212 pKa = 3.93FNIGSKK218 pKa = 9.99YY219 pKa = 10.35RR220 pKa = 11.84EE221 pKa = 4.16INVTDD226 pKa = 4.14PRR228 pKa = 11.84DD229 pKa = 3.56GDD231 pKa = 3.53ILRR234 pKa = 11.84AFNNFSVLMFRR245 pKa = 11.84FSYY248 pKa = 10.67RR249 pKa = 11.84LDD251 pKa = 3.23FRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84TLRR258 pKa = 11.84PNAIGFHH265 pKa = 5.38TKK267 pKa = 8.6KK268 pKa = 10.28QKK270 pKa = 10.25PIPHH274 pKa = 6.2SEE276 pKa = 4.1EE277 pKa = 3.9SHH279 pKa = 5.65TSII282 pKa = 5.37
Molecular weight: 33.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1614774 |
39 |
5059 |
342.5 |
38.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.114 ± 0.032 | 0.745 ± 0.012 |
5.816 ± 0.034 | 6.633 ± 0.043 |
5.226 ± 0.03 | 6.492 ± 0.043 |
1.837 ± 0.017 | 7.889 ± 0.032 |
7.348 ± 0.056 | 9.097 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.019 | 6.009 ± 0.037 |
3.447 ± 0.027 | 3.73 ± 0.023 |
3.593 ± 0.024 | 6.473 ± 0.029 |
6.016 ± 0.047 | 6.24 ± 0.027 |
1.101 ± 0.014 | 4.12 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
