
Alteromonas phage JH01
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
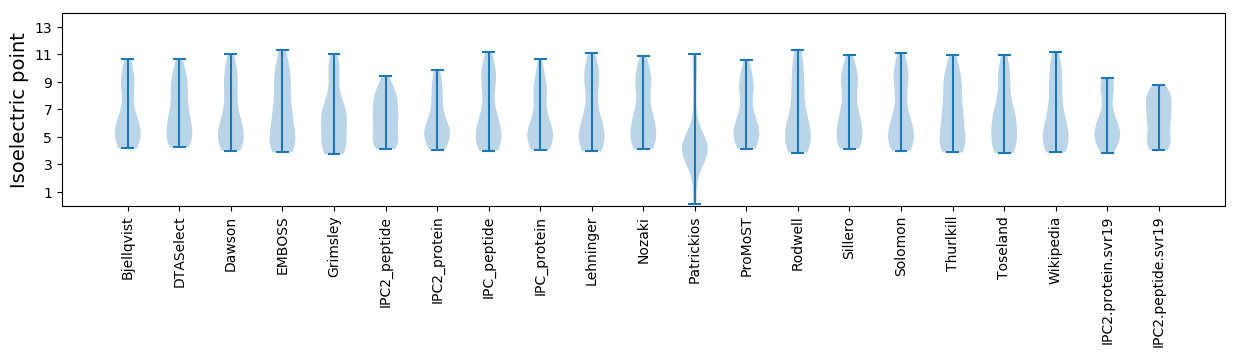
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2Z4PYC9|A0A2Z4PYC9_9CAUD Uncharacterized protein OS=Alteromonas phage JH01 OX=2249314 PE=4 SV=1
MM1 pKa = 7.89PFLISGTITNNGNPVSVPVVAFSTTVPSILLGSTVSAADD40 pKa = 3.36GKK42 pKa = 8.86YY43 pKa = 9.03TISEE47 pKa = 4.26LEE49 pKa = 4.17YY50 pKa = 9.65KK51 pKa = 10.84GSAIVVAYY59 pKa = 10.84SNGGTLFNPTTVVLTDD75 pKa = 5.02DD76 pKa = 4.86IISPVPANGYY86 pKa = 8.18VYY88 pKa = 10.63RR89 pKa = 11.84ALNPGTLGNVEE100 pKa = 4.76PEE102 pKa = 3.8WATDD106 pKa = 3.49NNVISGDD113 pKa = 3.55VTLEE117 pKa = 4.39PIQVFAPSAAGYY129 pKa = 9.28EE130 pKa = 4.08NTLFYY135 pKa = 10.78MPDD138 pKa = 2.88SVQYY142 pKa = 10.39KK143 pKa = 9.41GRR145 pKa = 11.84NLVRR149 pKa = 11.84GTSAVVGGKK158 pKa = 7.9TYY160 pKa = 10.83YY161 pKa = 10.66AYY163 pKa = 11.06SDD165 pKa = 4.07TNDD168 pKa = 3.37PTYY171 pKa = 10.5IGYY174 pKa = 9.82FDD176 pKa = 3.91EE177 pKa = 5.71LIRR180 pKa = 11.84PLSITQLNVVNADD193 pKa = 3.7FEE195 pKa = 4.72TGDD198 pKa = 3.62LSNWQTTKK206 pKa = 9.59GTPDD210 pKa = 2.88VRR212 pKa = 11.84EE213 pKa = 3.83IARR216 pKa = 11.84GDD218 pKa = 3.46DD219 pKa = 3.42VRR221 pKa = 11.84NVVYY225 pKa = 10.27GAATLEE231 pKa = 4.31WGVSQQISLPILLSQIGYY249 pKa = 6.68PTDD252 pKa = 5.08AIPFLFDD259 pKa = 3.39NVTITLKK266 pKa = 10.54VAQSSYY272 pKa = 11.09NNSDD276 pKa = 3.37PAQQYY281 pKa = 9.94IRR283 pKa = 11.84LIDD286 pKa = 3.86SNNSEE291 pKa = 4.51ISFEE295 pKa = 4.51SNDD298 pKa = 3.67LKK300 pKa = 10.49TEE302 pKa = 4.14SSLEE306 pKa = 3.95WVEE309 pKa = 4.02RR310 pKa = 11.84TVSISANSNMDD321 pKa = 3.59LNAIEE326 pKa = 4.29VEE328 pKa = 4.25LYY330 pKa = 10.46GYY332 pKa = 7.65RR333 pKa = 11.84TNGSNNDD340 pKa = 3.36GYY342 pKa = 7.46TTSVRR347 pKa = 11.84VEE349 pKa = 4.19VVANEE354 pKa = 3.82
MM1 pKa = 7.89PFLISGTITNNGNPVSVPVVAFSTTVPSILLGSTVSAADD40 pKa = 3.36GKK42 pKa = 8.86YY43 pKa = 9.03TISEE47 pKa = 4.26LEE49 pKa = 4.17YY50 pKa = 9.65KK51 pKa = 10.84GSAIVVAYY59 pKa = 10.84SNGGTLFNPTTVVLTDD75 pKa = 5.02DD76 pKa = 4.86IISPVPANGYY86 pKa = 8.18VYY88 pKa = 10.63RR89 pKa = 11.84ALNPGTLGNVEE100 pKa = 4.76PEE102 pKa = 3.8WATDD106 pKa = 3.49NNVISGDD113 pKa = 3.55VTLEE117 pKa = 4.39PIQVFAPSAAGYY129 pKa = 9.28EE130 pKa = 4.08NTLFYY135 pKa = 10.78MPDD138 pKa = 2.88SVQYY142 pKa = 10.39KK143 pKa = 9.41GRR145 pKa = 11.84NLVRR149 pKa = 11.84GTSAVVGGKK158 pKa = 7.9TYY160 pKa = 10.83YY161 pKa = 10.66AYY163 pKa = 11.06SDD165 pKa = 4.07TNDD168 pKa = 3.37PTYY171 pKa = 10.5IGYY174 pKa = 9.82FDD176 pKa = 3.91EE177 pKa = 5.71LIRR180 pKa = 11.84PLSITQLNVVNADD193 pKa = 3.7FEE195 pKa = 4.72TGDD198 pKa = 3.62LSNWQTTKK206 pKa = 9.59GTPDD210 pKa = 2.88VRR212 pKa = 11.84EE213 pKa = 3.83IARR216 pKa = 11.84GDD218 pKa = 3.46DD219 pKa = 3.42VRR221 pKa = 11.84NVVYY225 pKa = 10.27GAATLEE231 pKa = 4.31WGVSQQISLPILLSQIGYY249 pKa = 6.68PTDD252 pKa = 5.08AIPFLFDD259 pKa = 3.39NVTITLKK266 pKa = 10.54VAQSSYY272 pKa = 11.09NNSDD276 pKa = 3.37PAQQYY281 pKa = 9.94IRR283 pKa = 11.84LIDD286 pKa = 3.86SNNSEE291 pKa = 4.51ISFEE295 pKa = 4.51SNDD298 pKa = 3.67LKK300 pKa = 10.49TEE302 pKa = 4.14SSLEE306 pKa = 3.95WVEE309 pKa = 4.02RR310 pKa = 11.84TVSISANSNMDD321 pKa = 3.59LNAIEE326 pKa = 4.29VEE328 pKa = 4.25LYY330 pKa = 10.46GYY332 pKa = 7.65RR333 pKa = 11.84TNGSNNDD340 pKa = 3.36GYY342 pKa = 7.46TTSVRR347 pKa = 11.84VEE349 pKa = 4.19VVANEE354 pKa = 3.82
Molecular weight: 38.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4Q0C9|A0A2Z4Q0C9_9CAUD Uncharacterized protein OS=Alteromonas phage JH01 OX=2249314 PE=4 SV=1
MM1 pKa = 7.68AKK3 pKa = 9.13FTAEE7 pKa = 3.85QLGVLAEE14 pKa = 4.38KK15 pKa = 10.42IKK17 pKa = 11.29ANNTTINRR25 pKa = 11.84AASLAINKK33 pKa = 7.5TVTYY37 pKa = 10.39AKK39 pKa = 10.02DD40 pKa = 3.43LSISEE45 pKa = 3.95IVRR48 pKa = 11.84NVNLQQSYY56 pKa = 10.55IKK58 pKa = 10.62QNLKK62 pKa = 6.17TAKK65 pKa = 9.7RR66 pKa = 11.84ASPSDD71 pKa = 3.26LSAIIRR77 pKa = 11.84ANTRR81 pKa = 11.84EE82 pKa = 4.14TLLTRR87 pKa = 11.84YY88 pKa = 9.03PYY90 pKa = 10.47QEE92 pKa = 3.8TSTGVRR98 pKa = 11.84VAINKK103 pKa = 5.54TTGYY107 pKa = 8.72RR108 pKa = 11.84TINRR112 pKa = 11.84AFRR115 pKa = 11.84VTNLRR120 pKa = 11.84GSGATGIALKK130 pKa = 10.88NKK132 pKa = 9.91DD133 pKa = 3.26AVEE136 pKa = 4.02VFRR139 pKa = 11.84RR140 pKa = 11.84SLSPSTPGKK149 pKa = 9.54AAKK152 pKa = 9.71LARR155 pKa = 11.84IEE157 pKa = 4.6AKK159 pKa = 10.81AEE161 pKa = 4.08TKK163 pKa = 10.6PNGITVLHH171 pKa = 6.32SRR173 pKa = 11.84SINQLFTSVRR183 pKa = 11.84EE184 pKa = 4.08DD185 pKa = 3.43VQPRR189 pKa = 11.84TYY191 pKa = 11.03RR192 pKa = 11.84FMADD196 pKa = 3.13QFISDD201 pKa = 4.18LQRR204 pKa = 11.84LKK206 pKa = 11.18GKK208 pKa = 9.55SS209 pKa = 3.14
MM1 pKa = 7.68AKK3 pKa = 9.13FTAEE7 pKa = 3.85QLGVLAEE14 pKa = 4.38KK15 pKa = 10.42IKK17 pKa = 11.29ANNTTINRR25 pKa = 11.84AASLAINKK33 pKa = 7.5TVTYY37 pKa = 10.39AKK39 pKa = 10.02DD40 pKa = 3.43LSISEE45 pKa = 3.95IVRR48 pKa = 11.84NVNLQQSYY56 pKa = 10.55IKK58 pKa = 10.62QNLKK62 pKa = 6.17TAKK65 pKa = 9.7RR66 pKa = 11.84ASPSDD71 pKa = 3.26LSAIIRR77 pKa = 11.84ANTRR81 pKa = 11.84EE82 pKa = 4.14TLLTRR87 pKa = 11.84YY88 pKa = 9.03PYY90 pKa = 10.47QEE92 pKa = 3.8TSTGVRR98 pKa = 11.84VAINKK103 pKa = 5.54TTGYY107 pKa = 8.72RR108 pKa = 11.84TINRR112 pKa = 11.84AFRR115 pKa = 11.84VTNLRR120 pKa = 11.84GSGATGIALKK130 pKa = 10.88NKK132 pKa = 9.91DD133 pKa = 3.26AVEE136 pKa = 4.02VFRR139 pKa = 11.84RR140 pKa = 11.84SLSPSTPGKK149 pKa = 9.54AAKK152 pKa = 9.71LARR155 pKa = 11.84IEE157 pKa = 4.6AKK159 pKa = 10.81AEE161 pKa = 4.08TKK163 pKa = 10.6PNGITVLHH171 pKa = 6.32SRR173 pKa = 11.84SINQLFTSVRR183 pKa = 11.84EE184 pKa = 4.08DD185 pKa = 3.43VQPRR189 pKa = 11.84TYY191 pKa = 11.03RR192 pKa = 11.84FMADD196 pKa = 3.13QFISDD201 pKa = 4.18LQRR204 pKa = 11.84LKK206 pKa = 11.18GKK208 pKa = 9.55SS209 pKa = 3.14
Molecular weight: 23.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14468 |
39 |
1400 |
249.4 |
27.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.412 ± 0.66 | 1.092 ± 0.189 |
6.207 ± 0.277 | 6.919 ± 0.514 |
3.49 ± 0.24 | 6.359 ± 0.276 |
1.825 ± 0.286 | 5.391 ± 0.191 |
5.426 ± 0.387 | 8.301 ± 0.584 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.046 ± 0.229 | 5.391 ± 0.302 |
4.036 ± 0.482 | 4.147 ± 0.526 |
5.267 ± 0.322 | 6.186 ± 0.356 |
7.071 ± 0.357 | 7.7 ± 0.476 |
1.127 ± 0.14 | 3.608 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
