
Trinickia fusca
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Trinickia
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
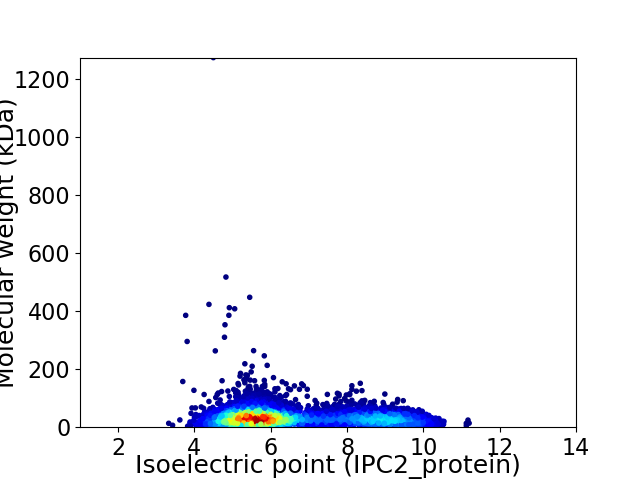
Virtual 2D-PAGE plot for 5256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A494XE96|A0A494XE96_9BURK Type II secretion system protein OS=Trinickia fusca OX=2419777 GN=D7S89_17740 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.04FAARR6 pKa = 11.84LLKK9 pKa = 10.78YY10 pKa = 9.92FAARR14 pKa = 11.84PAPRR18 pKa = 11.84VTAVAPAPLLLALEE32 pKa = 4.11PRR34 pKa = 11.84IVYY37 pKa = 9.16DD38 pKa = 3.56ASVAAVGAAARR49 pKa = 11.84HH50 pKa = 5.03HH51 pKa = 6.69VEE53 pKa = 4.16PGADD57 pKa = 3.03AAHH60 pKa = 7.31RR61 pKa = 11.84IGEE64 pKa = 4.32AVAHH68 pKa = 6.11GRR70 pKa = 11.84PAQNDD75 pKa = 3.75SVTPANGGADD85 pKa = 2.89RR86 pKa = 11.84HH87 pKa = 5.69TNQANPAQRR96 pKa = 11.84NVMADD101 pKa = 3.3AVQGSTQVVFVDD113 pKa = 4.05PAVTGYY119 pKa = 7.94QTLLAGLPAGTQVVMLNANTDD140 pKa = 3.09GFAQIAQYY148 pKa = 10.01LQQHH152 pKa = 6.62PGVSAIHH159 pKa = 6.18LLSHH163 pKa = 6.4GAEE166 pKa = 4.14GDD168 pKa = 3.94VQAGSAWLDD177 pKa = 3.15PANFSKK183 pKa = 11.11YY184 pKa = 9.54SAEE187 pKa = 3.94LAQIGAAMQPGGDD200 pKa = 3.81FLIYY204 pKa = 10.54GCDD207 pKa = 3.36VAEE210 pKa = 4.56GADD213 pKa = 3.54GRR215 pKa = 11.84LLVQEE220 pKa = 4.68IAAATHH226 pKa = 6.24LNVAASTDD234 pKa = 3.67LTGAASLGGNWTLEE248 pKa = 4.1YY249 pKa = 10.57QVGNVQTPVIFSAAAEE265 pKa = 4.04QQYY268 pKa = 11.21GSLLSEE274 pKa = 4.42TIEE277 pKa = 5.11DD278 pKa = 3.95YY279 pKa = 11.1TSAAASSLNTGDD291 pKa = 3.45VTSFSLDD298 pKa = 3.64GITYY302 pKa = 8.08TYY304 pKa = 11.23NVANATTVGPDD315 pKa = 3.3SNLQSLPDD323 pKa = 3.7EE324 pKa = 4.82NGSSEE329 pKa = 4.42SLVVNQDD336 pKa = 3.0GVSGLTSITITMQDD350 pKa = 2.51GKK352 pKa = 10.56AFRR355 pKa = 11.84MSSFDD360 pKa = 3.29IDD362 pKa = 3.22IVTNTVATIEE372 pKa = 4.5ANGSTLNDD380 pKa = 3.01ITITSNGVFQTEE392 pKa = 4.53TVDD395 pKa = 3.57LTGDD399 pKa = 3.56SAFDD403 pKa = 3.46VTNSITITFQSNSIVNLGHH422 pKa = 7.55LVYY425 pKa = 10.73QEE427 pKa = 3.8VTPPVVTATGGATTFTSVDD446 pKa = 3.3SSTATPVAVDD456 pKa = 3.2SGVTVSDD463 pKa = 3.99TEE465 pKa = 4.4SGTMSSATVAITGNFHH481 pKa = 7.17SGQDD485 pKa = 3.39VLAFTNTSSVIYY497 pKa = 9.99GSISGSYY504 pKa = 9.73NATTGVLTLSGAGTTAQYY522 pKa = 10.5QAALEE527 pKa = 4.17AVTYY531 pKa = 10.51LDD533 pKa = 3.63TAGTPNTSARR543 pKa = 11.84TISFSVNDD551 pKa = 5.7GIASSTVVTKK561 pKa = 10.15TVDD564 pKa = 3.04VDD566 pKa = 3.91SPPVVTTTGGTTNYY580 pKa = 10.51YY581 pKa = 10.77GGTSATAVDD590 pKa = 4.04SGVSVTDD597 pKa = 4.08ASQGTQASGTVTITSGYY614 pKa = 10.51DD615 pKa = 3.16SSNDD619 pKa = 3.04TLAFTNTSLTLFGNISASFNSSTGVLTLSSSGSTATDD656 pKa = 3.53AQWANAFEE664 pKa = 4.63AVMFSSASTTYY675 pKa = 10.93GNRR678 pKa = 11.84TISFVVNDD686 pKa = 3.93GTDD689 pKa = 3.15NSVAATKK696 pKa = 10.28IVDD699 pKa = 3.82VLNPSPVVTADD710 pKa = 3.09SGSAAFTAGDD720 pKa = 3.87NTTSTPVAIDD730 pKa = 3.14SGLTVTDD737 pKa = 4.62GNSSTLASATIAITGNFHH755 pKa = 7.25SGQDD759 pKa = 3.48VLAFTNNGSTMGNIAGAYY777 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD803 pKa = 3.76AVTYY807 pKa = 9.17TDD809 pKa = 4.21TAITPNNATRR819 pKa = 11.84TISFTASDD827 pKa = 4.24GVNTSNTATRR837 pKa = 11.84TVTVADD843 pKa = 3.69VDD845 pKa = 3.57QTPIVTTTGGTTNYY859 pKa = 10.29VGGASATTVDD869 pKa = 3.56GGVTVSDD876 pKa = 4.91LDD878 pKa = 3.79NATQASGTVSIGSGFHH894 pKa = 7.01SGDD897 pKa = 3.19TLSFTNTSLTLFGNISASYY916 pKa = 10.18NSGTGVLTLSSSGATATDD934 pKa = 3.77AQWANAFDD942 pKa = 4.16AVTFSSTSTTYY953 pKa = 11.28GNRR956 pKa = 11.84TVSFVVNDD964 pKa = 3.78GTEE967 pKa = 3.73NSAAATKK974 pKa = 9.63TVDD977 pKa = 3.84VINPNPVVTTDD988 pKa = 3.04SGSAAFTAGDD998 pKa = 3.87NTTSTPVVIDD1008 pKa = 3.29SGLTVTDD1015 pKa = 4.62GNSSTLASATIAITGNFHH1033 pKa = 7.25SGQDD1037 pKa = 3.49VLAFTNDD1044 pKa = 2.85GSTMGNIAGAYY1055 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD1081 pKa = 4.0SVTYY1085 pKa = 9.91TDD1087 pKa = 4.2TAVTPNNATRR1097 pKa = 11.84TISFTASDD1105 pKa = 4.22GVNTSSTATRR1115 pKa = 11.84TVTVADD1121 pKa = 3.69VDD1123 pKa = 3.57QTPIVTTTGGTTTYY1137 pKa = 9.91TAGASATTVDD1147 pKa = 3.56GGVTVSDD1154 pKa = 5.02LDD1156 pKa = 3.66NTTQASGTVTIGSGFHH1172 pKa = 6.89SGDD1175 pKa = 3.19TLSFTNTSLTLFGNIVAVYY1194 pKa = 10.14NSGTGVLTLSSSGASATDD1212 pKa = 3.68AQWANAFDD1220 pKa = 4.26AVTFSAGASATTGNRR1235 pKa = 11.84TISFVVNDD1243 pKa = 3.9GTEE1246 pKa = 3.73NSAAATKK1253 pKa = 9.67TVDD1256 pKa = 3.31VLGPPTVTTDD1266 pKa = 2.89SGSAAFTAGDD1276 pKa = 3.9NVTSTPVTIDD1286 pKa = 2.77SGLTVTDD1293 pKa = 5.04GSSSTLASATIAITGNFHH1311 pKa = 7.25SGQDD1315 pKa = 3.49VLAFTNDD1322 pKa = 2.85GSTMGNIAGAYY1333 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD1359 pKa = 3.76AVTYY1363 pKa = 9.81TDD1365 pKa = 3.91TAVTPNNATRR1375 pKa = 11.84TISFTTNDD1383 pKa = 3.57GVNTSNTATRR1393 pKa = 11.84TVTVADD1399 pKa = 3.69VDD1401 pKa = 3.57QTPIVTTTGGTTNYY1415 pKa = 10.33VGGTSATAVDD1425 pKa = 3.91GGVTVSDD1432 pKa = 4.34RR1433 pKa = 11.84DD1434 pKa = 3.56NTTQSSGTVSIGSGFHH1450 pKa = 7.01SGDD1453 pKa = 3.19TLSFTNTSSLLFGNIVASYY1472 pKa = 10.05NAGTGVLTLSSSGASATDD1490 pKa = 3.68AQWANAFDD1498 pKa = 4.26AVTFSAGASATTGNRR1513 pKa = 11.84TISFVVNDD1521 pKa = 3.9GTEE1524 pKa = 3.73NSAAATKK1531 pKa = 9.67TVDD1534 pKa = 3.31VLGPPTVTTDD1544 pKa = 2.89SGSAAFTAGDD1554 pKa = 3.9NVTSTPVVIDD1564 pKa = 3.29SGLTVTDD1571 pKa = 5.15GSSSTT1576 pKa = 3.75
MM1 pKa = 7.35KK2 pKa = 10.04FAARR6 pKa = 11.84LLKK9 pKa = 10.78YY10 pKa = 9.92FAARR14 pKa = 11.84PAPRR18 pKa = 11.84VTAVAPAPLLLALEE32 pKa = 4.11PRR34 pKa = 11.84IVYY37 pKa = 9.16DD38 pKa = 3.56ASVAAVGAAARR49 pKa = 11.84HH50 pKa = 5.03HH51 pKa = 6.69VEE53 pKa = 4.16PGADD57 pKa = 3.03AAHH60 pKa = 7.31RR61 pKa = 11.84IGEE64 pKa = 4.32AVAHH68 pKa = 6.11GRR70 pKa = 11.84PAQNDD75 pKa = 3.75SVTPANGGADD85 pKa = 2.89RR86 pKa = 11.84HH87 pKa = 5.69TNQANPAQRR96 pKa = 11.84NVMADD101 pKa = 3.3AVQGSTQVVFVDD113 pKa = 4.05PAVTGYY119 pKa = 7.94QTLLAGLPAGTQVVMLNANTDD140 pKa = 3.09GFAQIAQYY148 pKa = 10.01LQQHH152 pKa = 6.62PGVSAIHH159 pKa = 6.18LLSHH163 pKa = 6.4GAEE166 pKa = 4.14GDD168 pKa = 3.94VQAGSAWLDD177 pKa = 3.15PANFSKK183 pKa = 11.11YY184 pKa = 9.54SAEE187 pKa = 3.94LAQIGAAMQPGGDD200 pKa = 3.81FLIYY204 pKa = 10.54GCDD207 pKa = 3.36VAEE210 pKa = 4.56GADD213 pKa = 3.54GRR215 pKa = 11.84LLVQEE220 pKa = 4.68IAAATHH226 pKa = 6.24LNVAASTDD234 pKa = 3.67LTGAASLGGNWTLEE248 pKa = 4.1YY249 pKa = 10.57QVGNVQTPVIFSAAAEE265 pKa = 4.04QQYY268 pKa = 11.21GSLLSEE274 pKa = 4.42TIEE277 pKa = 5.11DD278 pKa = 3.95YY279 pKa = 11.1TSAAASSLNTGDD291 pKa = 3.45VTSFSLDD298 pKa = 3.64GITYY302 pKa = 8.08TYY304 pKa = 11.23NVANATTVGPDD315 pKa = 3.3SNLQSLPDD323 pKa = 3.7EE324 pKa = 4.82NGSSEE329 pKa = 4.42SLVVNQDD336 pKa = 3.0GVSGLTSITITMQDD350 pKa = 2.51GKK352 pKa = 10.56AFRR355 pKa = 11.84MSSFDD360 pKa = 3.29IDD362 pKa = 3.22IVTNTVATIEE372 pKa = 4.5ANGSTLNDD380 pKa = 3.01ITITSNGVFQTEE392 pKa = 4.53TVDD395 pKa = 3.57LTGDD399 pKa = 3.56SAFDD403 pKa = 3.46VTNSITITFQSNSIVNLGHH422 pKa = 7.55LVYY425 pKa = 10.73QEE427 pKa = 3.8VTPPVVTATGGATTFTSVDD446 pKa = 3.3SSTATPVAVDD456 pKa = 3.2SGVTVSDD463 pKa = 3.99TEE465 pKa = 4.4SGTMSSATVAITGNFHH481 pKa = 7.17SGQDD485 pKa = 3.39VLAFTNTSSVIYY497 pKa = 9.99GSISGSYY504 pKa = 9.73NATTGVLTLSGAGTTAQYY522 pKa = 10.5QAALEE527 pKa = 4.17AVTYY531 pKa = 10.51LDD533 pKa = 3.63TAGTPNTSARR543 pKa = 11.84TISFSVNDD551 pKa = 5.7GIASSTVVTKK561 pKa = 10.15TVDD564 pKa = 3.04VDD566 pKa = 3.91SPPVVTTTGGTTNYY580 pKa = 10.51YY581 pKa = 10.77GGTSATAVDD590 pKa = 4.04SGVSVTDD597 pKa = 4.08ASQGTQASGTVTITSGYY614 pKa = 10.51DD615 pKa = 3.16SSNDD619 pKa = 3.04TLAFTNTSLTLFGNISASFNSSTGVLTLSSSGSTATDD656 pKa = 3.53AQWANAFEE664 pKa = 4.63AVMFSSASTTYY675 pKa = 10.93GNRR678 pKa = 11.84TISFVVNDD686 pKa = 3.93GTDD689 pKa = 3.15NSVAATKK696 pKa = 10.28IVDD699 pKa = 3.82VLNPSPVVTADD710 pKa = 3.09SGSAAFTAGDD720 pKa = 3.87NTTSTPVAIDD730 pKa = 3.14SGLTVTDD737 pKa = 4.62GNSSTLASATIAITGNFHH755 pKa = 7.25SGQDD759 pKa = 3.48VLAFTNNGSTMGNIAGAYY777 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD803 pKa = 3.76AVTYY807 pKa = 9.17TDD809 pKa = 4.21TAITPNNATRR819 pKa = 11.84TISFTASDD827 pKa = 4.24GVNTSNTATRR837 pKa = 11.84TVTVADD843 pKa = 3.69VDD845 pKa = 3.57QTPIVTTTGGTTNYY859 pKa = 10.29VGGASATTVDD869 pKa = 3.56GGVTVSDD876 pKa = 4.91LDD878 pKa = 3.79NATQASGTVSIGSGFHH894 pKa = 7.01SGDD897 pKa = 3.19TLSFTNTSLTLFGNISASYY916 pKa = 10.18NSGTGVLTLSSSGATATDD934 pKa = 3.77AQWANAFDD942 pKa = 4.16AVTFSSTSTTYY953 pKa = 11.28GNRR956 pKa = 11.84TVSFVVNDD964 pKa = 3.78GTEE967 pKa = 3.73NSAAATKK974 pKa = 9.63TVDD977 pKa = 3.84VINPNPVVTTDD988 pKa = 3.04SGSAAFTAGDD998 pKa = 3.87NTTSTPVVIDD1008 pKa = 3.29SGLTVTDD1015 pKa = 4.62GNSSTLASATIAITGNFHH1033 pKa = 7.25SGQDD1037 pKa = 3.49VLAFTNDD1044 pKa = 2.85GSTMGNIAGAYY1055 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD1081 pKa = 4.0SVTYY1085 pKa = 9.91TDD1087 pKa = 4.2TAVTPNNATRR1097 pKa = 11.84TISFTASDD1105 pKa = 4.22GVNTSSTATRR1115 pKa = 11.84TVTVADD1121 pKa = 3.69VDD1123 pKa = 3.57QTPIVTTTGGTTTYY1137 pKa = 9.91TAGASATTVDD1147 pKa = 3.56GGVTVSDD1154 pKa = 5.02LDD1156 pKa = 3.66NTTQASGTVTIGSGFHH1172 pKa = 6.89SGDD1175 pKa = 3.19TLSFTNTSLTLFGNIVAVYY1194 pKa = 10.14NSGTGVLTLSSSGASATDD1212 pKa = 3.68AQWANAFDD1220 pKa = 4.26AVTFSAGASATTGNRR1235 pKa = 11.84TISFVVNDD1243 pKa = 3.9GTEE1246 pKa = 3.73NSAAATKK1253 pKa = 9.67TVDD1256 pKa = 3.31VLGPPTVTTDD1266 pKa = 2.89SGSAAFTAGDD1276 pKa = 3.9NVTSTPVTIDD1286 pKa = 2.77SGLTVTDD1293 pKa = 5.04GSSSTLASATIAITGNFHH1311 pKa = 7.25SGQDD1315 pKa = 3.49VLAFTNDD1322 pKa = 2.85GSTMGNIAGAYY1333 pKa = 7.7NATTGVLTLTSAGGIATLAQWQAALDD1359 pKa = 3.76AVTYY1363 pKa = 9.81TDD1365 pKa = 3.91TAVTPNNATRR1375 pKa = 11.84TISFTTNDD1383 pKa = 3.57GVNTSNTATRR1393 pKa = 11.84TVTVADD1399 pKa = 3.69VDD1401 pKa = 3.57QTPIVTTTGGTTNYY1415 pKa = 10.33VGGTSATAVDD1425 pKa = 3.91GGVTVSDD1432 pKa = 4.34RR1433 pKa = 11.84DD1434 pKa = 3.56NTTQSSGTVSIGSGFHH1450 pKa = 7.01SGDD1453 pKa = 3.19TLSFTNTSSLLFGNIVASYY1472 pKa = 10.05NAGTGVLTLSSSGASATDD1490 pKa = 3.68AQWANAFDD1498 pKa = 4.26AVTFSAGASATTGNRR1513 pKa = 11.84TISFVVNDD1521 pKa = 3.9GTEE1524 pKa = 3.73NSAAATKK1531 pKa = 9.67TVDD1534 pKa = 3.31VLGPPTVTTDD1544 pKa = 2.89SGSAAFTAGDD1554 pKa = 3.9NVTSTPVVIDD1564 pKa = 3.29SGLTVTDD1571 pKa = 5.15GSSSTT1576 pKa = 3.75
Molecular weight: 157.46 kDa
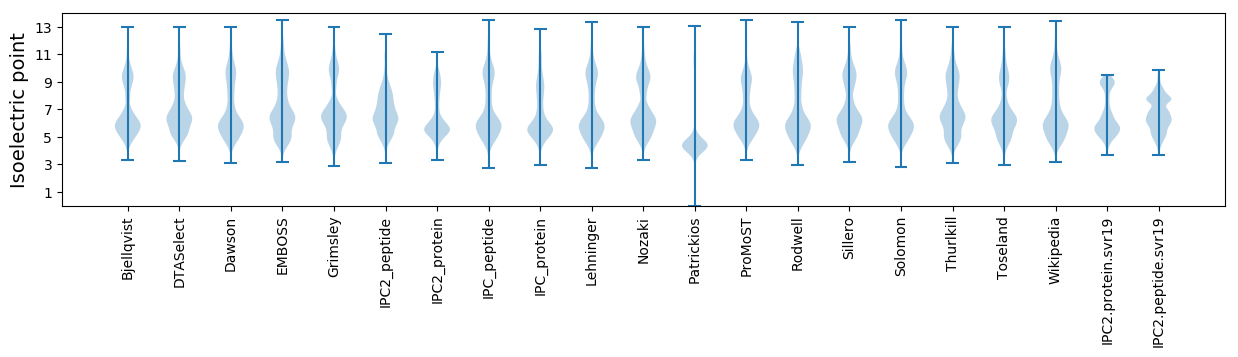
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A494XJQ5|A0A494XJQ5_9BURK Gluconokinase OS=Trinickia fusca OX=2419777 GN=D7S89_07835 PE=3 SV=1
MM1 pKa = 7.65LGGSAWPRR9 pKa = 11.84LAWLRR14 pKa = 11.84LAWLRR19 pKa = 11.84LVWLRR24 pKa = 11.84LVWLRR29 pKa = 11.84LVWLRR34 pKa = 11.84LVWLRR39 pKa = 11.84LVWLRR44 pKa = 11.84LVWLRR49 pKa = 11.84LVWLRR54 pKa = 11.84LAWPRR59 pKa = 11.84LAWPRR64 pKa = 11.84LAWPRR69 pKa = 11.84LAWLRR74 pKa = 11.84LARR77 pKa = 11.84PPLASHH83 pKa = 4.4QTVRR87 pKa = 11.84RR88 pKa = 11.84PLPSPHH94 pKa = 7.06PAWHH98 pKa = 6.59HH99 pKa = 5.97PPRR102 pKa = 11.84CPRR105 pKa = 11.84QRR107 pKa = 11.84LQPRR111 pKa = 11.84HH112 pKa = 5.18
MM1 pKa = 7.65LGGSAWPRR9 pKa = 11.84LAWLRR14 pKa = 11.84LAWLRR19 pKa = 11.84LVWLRR24 pKa = 11.84LVWLRR29 pKa = 11.84LVWLRR34 pKa = 11.84LVWLRR39 pKa = 11.84LVWLRR44 pKa = 11.84LVWLRR49 pKa = 11.84LVWLRR54 pKa = 11.84LAWPRR59 pKa = 11.84LAWPRR64 pKa = 11.84LAWPRR69 pKa = 11.84LAWLRR74 pKa = 11.84LARR77 pKa = 11.84PPLASHH83 pKa = 4.4QTVRR87 pKa = 11.84RR88 pKa = 11.84PLPSPHH94 pKa = 7.06PAWHH98 pKa = 6.59HH99 pKa = 5.97PPRR102 pKa = 11.84CPRR105 pKa = 11.84QRR107 pKa = 11.84LQPRR111 pKa = 11.84HH112 pKa = 5.18
Molecular weight: 13.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1758797 |
24 |
12197 |
334.6 |
36.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.914 ± 0.049 | 0.925 ± 0.013 |
5.44 ± 0.028 | 5.158 ± 0.044 |
3.552 ± 0.025 | 8.109 ± 0.053 |
2.367 ± 0.016 | 4.48 ± 0.024 |
2.911 ± 0.028 | 10.166 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.017 | 2.874 ± 0.043 |
5.06 ± 0.031 | 3.733 ± 0.027 |
6.887 ± 0.052 | 5.825 ± 0.037 |
5.756 ± 0.059 | 7.729 ± 0.025 |
1.359 ± 0.014 | 2.508 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
