
Pseudomonas fluorescens (strain F113)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas fluorescens group; Pseudomonas fluorescens
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
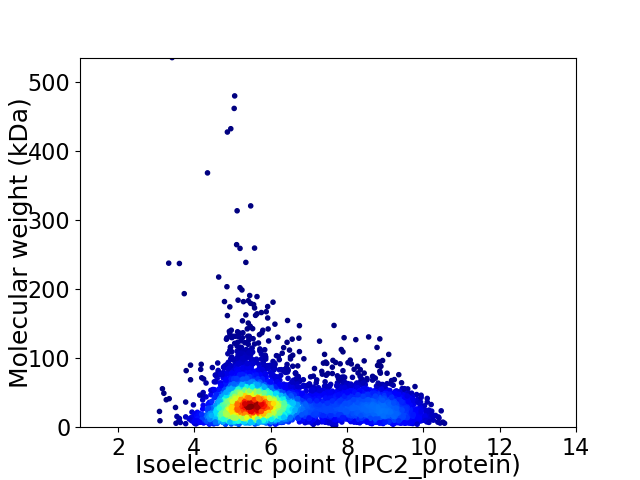
Virtual 2D-PAGE plot for 5837 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8PZR1|G8PZR1_PSEF3 Glyoxalase family protein superfamily OS=Pseudomonas fluorescens (strain F113) OX=1114970 GN=PSF113_2727 PE=4 SV=1
MM1 pKa = 7.56AALIGIVSKK10 pKa = 11.2VVGQVFAEE18 pKa = 4.09AAGGLRR24 pKa = 11.84RR25 pKa = 11.84PLVEE29 pKa = 4.19GDD31 pKa = 3.02RR32 pKa = 11.84LYY34 pKa = 11.13AGEE37 pKa = 4.25HH38 pKa = 5.9LVTGAEE44 pKa = 4.3GAVAVHH50 pKa = 5.68MQNGQTLTLGRR61 pKa = 11.84EE62 pKa = 4.36SNLTLTPQLLANHH75 pKa = 7.11APHH78 pKa = 7.02VDD80 pKa = 3.68TPDD83 pKa = 3.08AAAPSDD89 pKa = 3.73AQLTDD94 pKa = 3.49VQKK97 pKa = 10.75LQQAIAAGADD107 pKa = 3.42PTQTGEE113 pKa = 3.82ATAAGPEE120 pKa = 4.21GGNPGGVGGGHH131 pKa = 5.85SFVLLEE137 pKa = 4.15EE138 pKa = 4.42VGGEE142 pKa = 4.0VDD144 pKa = 3.89PLIGFPTAGFNGIPEE159 pKa = 4.33FPLLRR164 pKa = 11.84LAGDD168 pKa = 4.19PDD170 pKa = 3.72NSGDD174 pKa = 3.91DD175 pKa = 3.44ATVPPVTPEE184 pKa = 4.01TPDD187 pKa = 3.32NPVTLDD193 pKa = 3.44GVNVEE198 pKa = 4.27GGEE201 pKa = 4.18LTTNEE206 pKa = 4.18ANLADD211 pKa = 4.47GSASNPGALVQSGTFTVSAPDD232 pKa = 3.77GLSSLSIGGINVISGGVPAGFPQTITSALGNTLTITGYY270 pKa = 10.97DD271 pKa = 3.57PATGVVSYY279 pKa = 9.84TYY281 pKa = 10.63TLTDD285 pKa = 3.63NEE287 pKa = 4.34THH289 pKa = 6.8PAGGGANSITEE300 pKa = 4.14QFPVVAVDD308 pKa = 3.54TDD310 pKa = 3.44GDD312 pKa = 4.25TATGTLDD319 pKa = 3.87VNITDD324 pKa = 5.29DD325 pKa = 3.83VPQAIDD331 pKa = 3.76DD332 pKa = 4.04SHH334 pKa = 8.14ANTASEE340 pKa = 4.3TLVTLTGNVLPNDD353 pKa = 3.65HH354 pKa = 6.54QGADD358 pKa = 4.52RR359 pKa = 11.84IPTGPDD365 pKa = 2.56SGPIIGGTFTGTYY378 pKa = 7.22GTLVLNPNGTYY389 pKa = 9.73IYY391 pKa = 9.75TLNTSDD397 pKa = 3.65PQFVALHH404 pKa = 6.29GGGSGTEE411 pKa = 4.06TFTYY415 pKa = 10.31TLTDD419 pKa = 3.44ADD421 pKa = 4.77GDD423 pKa = 3.98TSTANLVLQVHH434 pKa = 6.33NNDD437 pKa = 3.98DD438 pKa = 3.89PVVLVGLDD446 pKa = 3.47AEE448 pKa = 4.66GGEE451 pKa = 5.36LSLQEE456 pKa = 4.79KK457 pKa = 10.07NLSDD461 pKa = 4.1GSNPDD466 pKa = 3.26ASALTQSGTFTVTALDD482 pKa = 4.08GVQTLSVGGINVVTGGVAAGFPQSITTALGNTLTITGFNAATGVVSYY529 pKa = 10.95SYY531 pKa = 10.27TLLDD535 pKa = 4.73NEE537 pKa = 4.59AHH539 pKa = 6.61PNANGANSVSEE550 pKa = 3.94QFAVVVTDD558 pKa = 5.35DD559 pKa = 4.31NGTTANGNLDD569 pKa = 3.66VNIVDD574 pKa = 5.11DD575 pKa = 4.5LPKK578 pKa = 10.85AVDD581 pKa = 4.02DD582 pKa = 4.9SNASTASEE590 pKa = 4.38TNLTLTGSVLTNDD603 pKa = 3.68TQGADD608 pKa = 3.54HH609 pKa = 6.49VASGPITPGTFTGTYY624 pKa = 7.23GTLVLNADD632 pKa = 3.56GSYY635 pKa = 10.19TYY637 pKa = 10.56TLNTADD643 pKa = 4.52ADD645 pKa = 4.28FKK647 pKa = 11.26GLHH650 pKa = 6.14GGGNGSEE657 pKa = 4.21TFTYY661 pKa = 10.51TLTDD665 pKa = 3.44ADD667 pKa = 4.77GDD669 pKa = 3.98TSTANLVLQVHH680 pKa = 6.32NNDD683 pKa = 3.95DD684 pKa = 3.82PVIITGLDD692 pKa = 3.43TEE694 pKa = 4.84GGEE697 pKa = 4.73LTIQEE702 pKa = 4.81KK703 pKa = 10.51NLSDD707 pKa = 3.9GSSPDD712 pKa = 3.37ASALTQSGTFIVTALDD728 pKa = 3.94GVQTLSVGGINVVTGGVAAGFPQSITTALGNTLTITGFNAATGVVSYY775 pKa = 10.95SYY777 pKa = 10.27TLLDD781 pKa = 4.73NEE783 pKa = 4.59AHH785 pKa = 6.61PNANGANSLSEE796 pKa = 3.87QFAVVVTDD804 pKa = 5.35DD805 pKa = 4.31NGTTANGNLDD815 pKa = 3.66VNIVDD820 pKa = 5.29DD821 pKa = 5.21LPTAHH826 pKa = 7.41ADD828 pKa = 3.44SASVDD833 pKa = 3.32EE834 pKa = 5.14GGTVSGNVLNNDD846 pKa = 3.19EE847 pKa = 4.87GGADD851 pKa = 3.72GPAASGAVIGVRR863 pKa = 11.84AGNDD867 pKa = 2.99TSTPAIGGLNTQINGTYY884 pKa = 10.63GYY886 pKa = 8.42LTLDD890 pKa = 3.37ANGNAVYY897 pKa = 10.14HH898 pKa = 6.15SNPNTVSAPGATDD911 pKa = 3.07VFTYY915 pKa = 7.69TVRR918 pKa = 11.84DD919 pKa = 3.44ADD921 pKa = 3.88GDD923 pKa = 3.89EE924 pKa = 4.27STTTITIDD932 pKa = 3.25VHH934 pKa = 6.52DD935 pKa = 4.63VCLVATPDD943 pKa = 3.57QEE945 pKa = 3.72ISVYY949 pKa = 10.56EE950 pKa = 4.0KK951 pKa = 11.09ALDD954 pKa = 4.02LNQDD958 pKa = 3.77GQDD961 pKa = 3.77LAPGTVTGSAPSATSEE977 pKa = 4.17TASGSLVGSISGAVGAVTFALVGNANGAYY1006 pKa = 9.61GQLSLQPDD1014 pKa = 3.98GSYY1017 pKa = 10.19TYY1019 pKa = 10.41TLTSPATTTPHH1030 pKa = 6.98ANDD1033 pKa = 3.96GPNVLSEE1040 pKa = 4.3SFTYY1044 pKa = 10.34QATDD1048 pKa = 3.04SLGNTVTSTIVIDD1061 pKa = 4.47IVDD1064 pKa = 4.47DD1065 pKa = 4.01VPNAHH1070 pKa = 7.28ADD1072 pKa = 3.52FASVVEE1078 pKa = 4.28GGTVNGNVLDD1088 pKa = 4.09NDD1090 pKa = 3.75VLGADD1095 pKa = 3.54GGMVVGVRR1103 pKa = 11.84AGNDD1107 pKa = 3.21TSNPAVGGLNTQVNGTYY1124 pKa = 10.69GYY1126 pKa = 8.5LTLDD1130 pKa = 3.37ANGNAVYY1137 pKa = 10.14HH1138 pKa = 6.15SNPNTVSAPGATDD1151 pKa = 3.07VFTYY1155 pKa = 7.69TVRR1158 pKa = 11.84DD1159 pKa = 3.44ADD1161 pKa = 3.88GDD1163 pKa = 3.89EE1164 pKa = 4.27STTTITIDD1172 pKa = 3.25VHH1174 pKa = 6.52DD1175 pKa = 4.63VCLVATPDD1183 pKa = 3.57QEE1185 pKa = 3.72ISVYY1189 pKa = 10.56EE1190 pKa = 4.0KK1191 pKa = 11.09ALDD1194 pKa = 4.02LNQDD1198 pKa = 3.77GQDD1201 pKa = 3.77LAPGTVTGSAPSATSEE1217 pKa = 4.17TASGSLVGSISGAVGAVTFALVGNANGAYY1246 pKa = 9.61GQLSLQPDD1254 pKa = 3.98GSYY1257 pKa = 10.19TYY1259 pKa = 10.41TLTSPATTTPHH1270 pKa = 6.98ANDD1273 pKa = 3.96GPNVLSEE1280 pKa = 4.3SFTYY1284 pKa = 10.34QATDD1288 pKa = 3.04SLGNTVTSTIVIDD1301 pKa = 4.47IVDD1304 pKa = 4.47DD1305 pKa = 4.01VPNAHH1310 pKa = 7.28ADD1312 pKa = 3.52FASVVEE1318 pKa = 4.28GGTVNGNVLDD1328 pKa = 4.09NDD1330 pKa = 3.75VLGADD1335 pKa = 3.6GGMVIGVRR1343 pKa = 11.84AGNDD1347 pKa = 3.2TSNPAVGGLNTQINGTYY1364 pKa = 10.63GYY1366 pKa = 8.42LTLDD1370 pKa = 3.37ANGNAVYY1377 pKa = 10.17HH1378 pKa = 6.18SNPDD1382 pKa = 3.18AVGAAGATDD1391 pKa = 3.31VFTYY1395 pKa = 7.69TVRR1398 pKa = 11.84DD1399 pKa = 3.44ADD1401 pKa = 3.88GDD1403 pKa = 3.89EE1404 pKa = 4.35STTTVTIDD1412 pKa = 3.11VHH1414 pKa = 6.09NSCLIVEE1421 pKa = 4.51TDD1423 pKa = 3.07HH1424 pKa = 7.88DD1425 pKa = 3.61ITVYY1429 pKa = 10.73EE1430 pKa = 4.43KK1431 pKa = 11.06ALDD1434 pKa = 4.09LNQDD1438 pKa = 3.77GQDD1441 pKa = 3.77LAPGTVTGSEE1451 pKa = 4.0PSATGEE1457 pKa = 4.13TASGSLVGSVSGATGAVTFTLVGNATGAYY1486 pKa = 8.53GQLLLHH1492 pKa = 7.18PDD1494 pKa = 3.23GSYY1497 pKa = 10.24TYY1499 pKa = 9.8TLTSPATTTPHH1510 pKa = 7.16ANDD1513 pKa = 4.01GPNALSEE1520 pKa = 4.45SFTYY1524 pKa = 10.33QATDD1528 pKa = 3.0ALGNSTTGSLVVSIVDD1544 pKa = 3.84DD1545 pKa = 3.93VPKK1548 pKa = 10.7AVASEE1553 pKa = 4.22RR1554 pKa = 11.84SVTAVEE1560 pKa = 4.07IDD1562 pKa = 3.66TNLLIVLDD1570 pKa = 4.28VSGSMKK1576 pKa = 10.53DD1577 pKa = 3.38DD1578 pKa = 3.81SGVSGLSRR1586 pKa = 11.84MEE1588 pKa = 4.09LAKK1591 pKa = 10.44QAISALLDD1599 pKa = 3.63KK1600 pKa = 10.72YY1601 pKa = 11.63DD1602 pKa = 4.35DD1603 pKa = 3.8MGDD1606 pKa = 3.42VKK1608 pKa = 11.12VQLVTFSGSATDD1620 pKa = 5.44QSTLWVDD1627 pKa = 3.28VATAKK1632 pKa = 9.99TLISALSAHH1641 pKa = 7.0GDD1643 pKa = 3.62TNYY1646 pKa = 11.0DD1647 pKa = 3.11AAVAVAKK1654 pKa = 9.21TAFANPGQLTGAQNVGYY1671 pKa = 9.32FFSDD1675 pKa = 3.77GKK1677 pKa = 8.61PTLGEE1682 pKa = 3.73IGSADD1687 pKa = 3.36EE1688 pKa = 4.51AAWKK1692 pKa = 10.66AFLDD1696 pKa = 3.71ANGIKK1701 pKa = 10.28NYY1703 pKa = 10.7AIGLGDD1709 pKa = 3.76GVSNTHH1715 pKa = 7.21LDD1717 pKa = 3.41PLAYY1721 pKa = 10.19DD1722 pKa = 4.67GSAHH1726 pKa = 6.29TNTNAVVVTDD1736 pKa = 4.52LNQLNSVLSDD1746 pKa = 3.56TVQGAPVTGNLLGEE1760 pKa = 4.6GGSFGADD1767 pKa = 2.58GGFIKK1772 pKa = 10.79SLVVDD1777 pKa = 4.58GTTYY1781 pKa = 10.82SYY1783 pKa = 11.69DD1784 pKa = 3.28PAANGGHH1791 pKa = 6.42GALTASGGLNHH1802 pKa = 7.24GSFNTATNTLSIATDD1817 pKa = 3.31HH1818 pKa = 7.55DD1819 pKa = 3.72GTLVVNLDD1827 pKa = 3.49TGAFSYY1833 pKa = 10.3TSQTATSTLITEE1845 pKa = 4.94HH1846 pKa = 6.71IGYY1849 pKa = 7.54TVSDD1853 pKa = 3.81NDD1855 pKa = 3.87GDD1857 pKa = 4.17LASSNLVINVVPNSPPIAVDD1877 pKa = 3.84DD1878 pKa = 4.75NIITNVLSGNIVIPGEE1894 pKa = 4.02LLLGNDD1900 pKa = 3.64SDD1902 pKa = 5.05VNGDD1906 pKa = 4.27PLTASPTSFNTGWTAKK1922 pKa = 10.54GGDD1925 pKa = 3.87FTGSTGTASFTGNNVQSINLSRR1947 pKa = 11.84SAFVANTASMTAMLVVSGALGMVSNNNANDD1977 pKa = 3.63EE1978 pKa = 4.06DD1979 pKa = 4.19RR1980 pKa = 11.84LNISLKK1986 pKa = 10.4QGEE1989 pKa = 5.02TLTLDD1994 pKa = 3.72HH1995 pKa = 6.73NLSAGRR2001 pKa = 11.84IAMEE2005 pKa = 3.81YY2006 pKa = 9.91SLNGGPFIAINDD2018 pKa = 4.27GASFTAAADD2027 pKa = 3.55GNYY2030 pKa = 9.4QIHH2033 pKa = 6.96VINIANPGGGNSNAAEE2049 pKa = 4.24NYY2051 pKa = 9.88QLILTVDD2058 pKa = 3.37YY2059 pKa = 11.39AGAQDD2064 pKa = 4.01KK2065 pKa = 9.94TPDD2068 pKa = 3.34YY2069 pKa = 9.81HH2070 pKa = 8.77GSYY2073 pKa = 8.77TASDD2077 pKa = 3.33NHH2079 pKa = 6.58GGSDD2083 pKa = 3.55SASVDD2088 pKa = 2.75ISYY2091 pKa = 10.68QAGHH2095 pKa = 6.53TLTGTSGDD2103 pKa = 3.88DD2104 pKa = 3.28VLVAGNGDD2112 pKa = 3.59NLLNAGDD2119 pKa = 4.26GNDD2122 pKa = 3.46ILSAGSGNNTLHH2134 pKa = 7.02GEE2136 pKa = 4.19AGDD2139 pKa = 3.96DD2140 pKa = 3.91LLYY2143 pKa = 11.17SGAGNDD2149 pKa = 4.36LLDD2152 pKa = 4.53GGTGNDD2158 pKa = 3.47TASYY2162 pKa = 10.05AHH2164 pKa = 6.97ATTGVTVNLGLLAAQNTLGAGTDD2187 pKa = 3.46TLAGIEE2193 pKa = 4.14NLMGSNFNDD2202 pKa = 3.63TLIGDD2207 pKa = 4.45GASNRR2212 pKa = 11.84INGGLGHH2219 pKa = 7.31DD2220 pKa = 4.01VLNGGGGDD2228 pKa = 3.91DD2229 pKa = 4.69LLIGGLGNNTLTGGSGADD2247 pKa = 3.36TFQWQAGNSGHH2258 pKa = 7.31DD2259 pKa = 4.0VITDD2263 pKa = 3.87FAPGVDD2269 pKa = 4.91KK2270 pKa = 11.41LDD2272 pKa = 4.61LSQLLQGEE2280 pKa = 4.45NGSAASLDD2288 pKa = 4.18DD2289 pKa = 3.73YY2290 pKa = 11.72LHH2292 pKa = 6.68FSVSGSGASVVTSIDD2307 pKa = 3.02VSAIAGATPNQTIDD2321 pKa = 3.45LTGVNLASHH2330 pKa = 6.36YY2331 pKa = 10.76GVTPGAGGIIGGADD2345 pKa = 3.06TATIINGMLNDD2356 pKa = 4.31HH2357 pKa = 6.08SLKK2360 pKa = 10.51VDD2362 pKa = 3.57TVV2364 pKa = 3.29
MM1 pKa = 7.56AALIGIVSKK10 pKa = 11.2VVGQVFAEE18 pKa = 4.09AAGGLRR24 pKa = 11.84RR25 pKa = 11.84PLVEE29 pKa = 4.19GDD31 pKa = 3.02RR32 pKa = 11.84LYY34 pKa = 11.13AGEE37 pKa = 4.25HH38 pKa = 5.9LVTGAEE44 pKa = 4.3GAVAVHH50 pKa = 5.68MQNGQTLTLGRR61 pKa = 11.84EE62 pKa = 4.36SNLTLTPQLLANHH75 pKa = 7.11APHH78 pKa = 7.02VDD80 pKa = 3.68TPDD83 pKa = 3.08AAAPSDD89 pKa = 3.73AQLTDD94 pKa = 3.49VQKK97 pKa = 10.75LQQAIAAGADD107 pKa = 3.42PTQTGEE113 pKa = 3.82ATAAGPEE120 pKa = 4.21GGNPGGVGGGHH131 pKa = 5.85SFVLLEE137 pKa = 4.15EE138 pKa = 4.42VGGEE142 pKa = 4.0VDD144 pKa = 3.89PLIGFPTAGFNGIPEE159 pKa = 4.33FPLLRR164 pKa = 11.84LAGDD168 pKa = 4.19PDD170 pKa = 3.72NSGDD174 pKa = 3.91DD175 pKa = 3.44ATVPPVTPEE184 pKa = 4.01TPDD187 pKa = 3.32NPVTLDD193 pKa = 3.44GVNVEE198 pKa = 4.27GGEE201 pKa = 4.18LTTNEE206 pKa = 4.18ANLADD211 pKa = 4.47GSASNPGALVQSGTFTVSAPDD232 pKa = 3.77GLSSLSIGGINVISGGVPAGFPQTITSALGNTLTITGYY270 pKa = 10.97DD271 pKa = 3.57PATGVVSYY279 pKa = 9.84TYY281 pKa = 10.63TLTDD285 pKa = 3.63NEE287 pKa = 4.34THH289 pKa = 6.8PAGGGANSITEE300 pKa = 4.14QFPVVAVDD308 pKa = 3.54TDD310 pKa = 3.44GDD312 pKa = 4.25TATGTLDD319 pKa = 3.87VNITDD324 pKa = 5.29DD325 pKa = 3.83VPQAIDD331 pKa = 3.76DD332 pKa = 4.04SHH334 pKa = 8.14ANTASEE340 pKa = 4.3TLVTLTGNVLPNDD353 pKa = 3.65HH354 pKa = 6.54QGADD358 pKa = 4.52RR359 pKa = 11.84IPTGPDD365 pKa = 2.56SGPIIGGTFTGTYY378 pKa = 7.22GTLVLNPNGTYY389 pKa = 9.73IYY391 pKa = 9.75TLNTSDD397 pKa = 3.65PQFVALHH404 pKa = 6.29GGGSGTEE411 pKa = 4.06TFTYY415 pKa = 10.31TLTDD419 pKa = 3.44ADD421 pKa = 4.77GDD423 pKa = 3.98TSTANLVLQVHH434 pKa = 6.33NNDD437 pKa = 3.98DD438 pKa = 3.89PVVLVGLDD446 pKa = 3.47AEE448 pKa = 4.66GGEE451 pKa = 5.36LSLQEE456 pKa = 4.79KK457 pKa = 10.07NLSDD461 pKa = 4.1GSNPDD466 pKa = 3.26ASALTQSGTFTVTALDD482 pKa = 4.08GVQTLSVGGINVVTGGVAAGFPQSITTALGNTLTITGFNAATGVVSYY529 pKa = 10.95SYY531 pKa = 10.27TLLDD535 pKa = 4.73NEE537 pKa = 4.59AHH539 pKa = 6.61PNANGANSVSEE550 pKa = 3.94QFAVVVTDD558 pKa = 5.35DD559 pKa = 4.31NGTTANGNLDD569 pKa = 3.66VNIVDD574 pKa = 5.11DD575 pKa = 4.5LPKK578 pKa = 10.85AVDD581 pKa = 4.02DD582 pKa = 4.9SNASTASEE590 pKa = 4.38TNLTLTGSVLTNDD603 pKa = 3.68TQGADD608 pKa = 3.54HH609 pKa = 6.49VASGPITPGTFTGTYY624 pKa = 7.23GTLVLNADD632 pKa = 3.56GSYY635 pKa = 10.19TYY637 pKa = 10.56TLNTADD643 pKa = 4.52ADD645 pKa = 4.28FKK647 pKa = 11.26GLHH650 pKa = 6.14GGGNGSEE657 pKa = 4.21TFTYY661 pKa = 10.51TLTDD665 pKa = 3.44ADD667 pKa = 4.77GDD669 pKa = 3.98TSTANLVLQVHH680 pKa = 6.32NNDD683 pKa = 3.95DD684 pKa = 3.82PVIITGLDD692 pKa = 3.43TEE694 pKa = 4.84GGEE697 pKa = 4.73LTIQEE702 pKa = 4.81KK703 pKa = 10.51NLSDD707 pKa = 3.9GSSPDD712 pKa = 3.37ASALTQSGTFIVTALDD728 pKa = 3.94GVQTLSVGGINVVTGGVAAGFPQSITTALGNTLTITGFNAATGVVSYY775 pKa = 10.95SYY777 pKa = 10.27TLLDD781 pKa = 4.73NEE783 pKa = 4.59AHH785 pKa = 6.61PNANGANSLSEE796 pKa = 3.87QFAVVVTDD804 pKa = 5.35DD805 pKa = 4.31NGTTANGNLDD815 pKa = 3.66VNIVDD820 pKa = 5.29DD821 pKa = 5.21LPTAHH826 pKa = 7.41ADD828 pKa = 3.44SASVDD833 pKa = 3.32EE834 pKa = 5.14GGTVSGNVLNNDD846 pKa = 3.19EE847 pKa = 4.87GGADD851 pKa = 3.72GPAASGAVIGVRR863 pKa = 11.84AGNDD867 pKa = 2.99TSTPAIGGLNTQINGTYY884 pKa = 10.63GYY886 pKa = 8.42LTLDD890 pKa = 3.37ANGNAVYY897 pKa = 10.14HH898 pKa = 6.15SNPNTVSAPGATDD911 pKa = 3.07VFTYY915 pKa = 7.69TVRR918 pKa = 11.84DD919 pKa = 3.44ADD921 pKa = 3.88GDD923 pKa = 3.89EE924 pKa = 4.27STTTITIDD932 pKa = 3.25VHH934 pKa = 6.52DD935 pKa = 4.63VCLVATPDD943 pKa = 3.57QEE945 pKa = 3.72ISVYY949 pKa = 10.56EE950 pKa = 4.0KK951 pKa = 11.09ALDD954 pKa = 4.02LNQDD958 pKa = 3.77GQDD961 pKa = 3.77LAPGTVTGSAPSATSEE977 pKa = 4.17TASGSLVGSISGAVGAVTFALVGNANGAYY1006 pKa = 9.61GQLSLQPDD1014 pKa = 3.98GSYY1017 pKa = 10.19TYY1019 pKa = 10.41TLTSPATTTPHH1030 pKa = 6.98ANDD1033 pKa = 3.96GPNVLSEE1040 pKa = 4.3SFTYY1044 pKa = 10.34QATDD1048 pKa = 3.04SLGNTVTSTIVIDD1061 pKa = 4.47IVDD1064 pKa = 4.47DD1065 pKa = 4.01VPNAHH1070 pKa = 7.28ADD1072 pKa = 3.52FASVVEE1078 pKa = 4.28GGTVNGNVLDD1088 pKa = 4.09NDD1090 pKa = 3.75VLGADD1095 pKa = 3.54GGMVVGVRR1103 pKa = 11.84AGNDD1107 pKa = 3.21TSNPAVGGLNTQVNGTYY1124 pKa = 10.69GYY1126 pKa = 8.5LTLDD1130 pKa = 3.37ANGNAVYY1137 pKa = 10.14HH1138 pKa = 6.15SNPNTVSAPGATDD1151 pKa = 3.07VFTYY1155 pKa = 7.69TVRR1158 pKa = 11.84DD1159 pKa = 3.44ADD1161 pKa = 3.88GDD1163 pKa = 3.89EE1164 pKa = 4.27STTTITIDD1172 pKa = 3.25VHH1174 pKa = 6.52DD1175 pKa = 4.63VCLVATPDD1183 pKa = 3.57QEE1185 pKa = 3.72ISVYY1189 pKa = 10.56EE1190 pKa = 4.0KK1191 pKa = 11.09ALDD1194 pKa = 4.02LNQDD1198 pKa = 3.77GQDD1201 pKa = 3.77LAPGTVTGSAPSATSEE1217 pKa = 4.17TASGSLVGSISGAVGAVTFALVGNANGAYY1246 pKa = 9.61GQLSLQPDD1254 pKa = 3.98GSYY1257 pKa = 10.19TYY1259 pKa = 10.41TLTSPATTTPHH1270 pKa = 6.98ANDD1273 pKa = 3.96GPNVLSEE1280 pKa = 4.3SFTYY1284 pKa = 10.34QATDD1288 pKa = 3.04SLGNTVTSTIVIDD1301 pKa = 4.47IVDD1304 pKa = 4.47DD1305 pKa = 4.01VPNAHH1310 pKa = 7.28ADD1312 pKa = 3.52FASVVEE1318 pKa = 4.28GGTVNGNVLDD1328 pKa = 4.09NDD1330 pKa = 3.75VLGADD1335 pKa = 3.6GGMVIGVRR1343 pKa = 11.84AGNDD1347 pKa = 3.2TSNPAVGGLNTQINGTYY1364 pKa = 10.63GYY1366 pKa = 8.42LTLDD1370 pKa = 3.37ANGNAVYY1377 pKa = 10.17HH1378 pKa = 6.18SNPDD1382 pKa = 3.18AVGAAGATDD1391 pKa = 3.31VFTYY1395 pKa = 7.69TVRR1398 pKa = 11.84DD1399 pKa = 3.44ADD1401 pKa = 3.88GDD1403 pKa = 3.89EE1404 pKa = 4.35STTTVTIDD1412 pKa = 3.11VHH1414 pKa = 6.09NSCLIVEE1421 pKa = 4.51TDD1423 pKa = 3.07HH1424 pKa = 7.88DD1425 pKa = 3.61ITVYY1429 pKa = 10.73EE1430 pKa = 4.43KK1431 pKa = 11.06ALDD1434 pKa = 4.09LNQDD1438 pKa = 3.77GQDD1441 pKa = 3.77LAPGTVTGSEE1451 pKa = 4.0PSATGEE1457 pKa = 4.13TASGSLVGSVSGATGAVTFTLVGNATGAYY1486 pKa = 8.53GQLLLHH1492 pKa = 7.18PDD1494 pKa = 3.23GSYY1497 pKa = 10.24TYY1499 pKa = 9.8TLTSPATTTPHH1510 pKa = 7.16ANDD1513 pKa = 4.01GPNALSEE1520 pKa = 4.45SFTYY1524 pKa = 10.33QATDD1528 pKa = 3.0ALGNSTTGSLVVSIVDD1544 pKa = 3.84DD1545 pKa = 3.93VPKK1548 pKa = 10.7AVASEE1553 pKa = 4.22RR1554 pKa = 11.84SVTAVEE1560 pKa = 4.07IDD1562 pKa = 3.66TNLLIVLDD1570 pKa = 4.28VSGSMKK1576 pKa = 10.53DD1577 pKa = 3.38DD1578 pKa = 3.81SGVSGLSRR1586 pKa = 11.84MEE1588 pKa = 4.09LAKK1591 pKa = 10.44QAISALLDD1599 pKa = 3.63KK1600 pKa = 10.72YY1601 pKa = 11.63DD1602 pKa = 4.35DD1603 pKa = 3.8MGDD1606 pKa = 3.42VKK1608 pKa = 11.12VQLVTFSGSATDD1620 pKa = 5.44QSTLWVDD1627 pKa = 3.28VATAKK1632 pKa = 9.99TLISALSAHH1641 pKa = 7.0GDD1643 pKa = 3.62TNYY1646 pKa = 11.0DD1647 pKa = 3.11AAVAVAKK1654 pKa = 9.21TAFANPGQLTGAQNVGYY1671 pKa = 9.32FFSDD1675 pKa = 3.77GKK1677 pKa = 8.61PTLGEE1682 pKa = 3.73IGSADD1687 pKa = 3.36EE1688 pKa = 4.51AAWKK1692 pKa = 10.66AFLDD1696 pKa = 3.71ANGIKK1701 pKa = 10.28NYY1703 pKa = 10.7AIGLGDD1709 pKa = 3.76GVSNTHH1715 pKa = 7.21LDD1717 pKa = 3.41PLAYY1721 pKa = 10.19DD1722 pKa = 4.67GSAHH1726 pKa = 6.29TNTNAVVVTDD1736 pKa = 4.52LNQLNSVLSDD1746 pKa = 3.56TVQGAPVTGNLLGEE1760 pKa = 4.6GGSFGADD1767 pKa = 2.58GGFIKK1772 pKa = 10.79SLVVDD1777 pKa = 4.58GTTYY1781 pKa = 10.82SYY1783 pKa = 11.69DD1784 pKa = 3.28PAANGGHH1791 pKa = 6.42GALTASGGLNHH1802 pKa = 7.24GSFNTATNTLSIATDD1817 pKa = 3.31HH1818 pKa = 7.55DD1819 pKa = 3.72GTLVVNLDD1827 pKa = 3.49TGAFSYY1833 pKa = 10.3TSQTATSTLITEE1845 pKa = 4.94HH1846 pKa = 6.71IGYY1849 pKa = 7.54TVSDD1853 pKa = 3.81NDD1855 pKa = 3.87GDD1857 pKa = 4.17LASSNLVINVVPNSPPIAVDD1877 pKa = 3.84DD1878 pKa = 4.75NIITNVLSGNIVIPGEE1894 pKa = 4.02LLLGNDD1900 pKa = 3.64SDD1902 pKa = 5.05VNGDD1906 pKa = 4.27PLTASPTSFNTGWTAKK1922 pKa = 10.54GGDD1925 pKa = 3.87FTGSTGTASFTGNNVQSINLSRR1947 pKa = 11.84SAFVANTASMTAMLVVSGALGMVSNNNANDD1977 pKa = 3.63EE1978 pKa = 4.06DD1979 pKa = 4.19RR1980 pKa = 11.84LNISLKK1986 pKa = 10.4QGEE1989 pKa = 5.02TLTLDD1994 pKa = 3.72HH1995 pKa = 6.73NLSAGRR2001 pKa = 11.84IAMEE2005 pKa = 3.81YY2006 pKa = 9.91SLNGGPFIAINDD2018 pKa = 4.27GASFTAAADD2027 pKa = 3.55GNYY2030 pKa = 9.4QIHH2033 pKa = 6.96VINIANPGGGNSNAAEE2049 pKa = 4.24NYY2051 pKa = 9.88QLILTVDD2058 pKa = 3.37YY2059 pKa = 11.39AGAQDD2064 pKa = 4.01KK2065 pKa = 9.94TPDD2068 pKa = 3.34YY2069 pKa = 9.81HH2070 pKa = 8.77GSYY2073 pKa = 8.77TASDD2077 pKa = 3.33NHH2079 pKa = 6.58GGSDD2083 pKa = 3.55SASVDD2088 pKa = 2.75ISYY2091 pKa = 10.68QAGHH2095 pKa = 6.53TLTGTSGDD2103 pKa = 3.88DD2104 pKa = 3.28VLVAGNGDD2112 pKa = 3.59NLLNAGDD2119 pKa = 4.26GNDD2122 pKa = 3.46ILSAGSGNNTLHH2134 pKa = 7.02GEE2136 pKa = 4.19AGDD2139 pKa = 3.96DD2140 pKa = 3.91LLYY2143 pKa = 11.17SGAGNDD2149 pKa = 4.36LLDD2152 pKa = 4.53GGTGNDD2158 pKa = 3.47TASYY2162 pKa = 10.05AHH2164 pKa = 6.97ATTGVTVNLGLLAAQNTLGAGTDD2187 pKa = 3.46TLAGIEE2193 pKa = 4.14NLMGSNFNDD2202 pKa = 3.63TLIGDD2207 pKa = 4.45GASNRR2212 pKa = 11.84INGGLGHH2219 pKa = 7.31DD2220 pKa = 4.01VLNGGGGDD2228 pKa = 3.91DD2229 pKa = 4.69LLIGGLGNNTLTGGSGADD2247 pKa = 3.36TFQWQAGNSGHH2258 pKa = 7.31DD2259 pKa = 4.0VITDD2263 pKa = 3.87FAPGVDD2269 pKa = 4.91KK2270 pKa = 11.41LDD2272 pKa = 4.61LSQLLQGEE2280 pKa = 4.45NGSAASLDD2288 pKa = 4.18DD2289 pKa = 3.73YY2290 pKa = 11.72LHH2292 pKa = 6.68FSVSGSGASVVTSIDD2307 pKa = 3.02VSAIAGATPNQTIDD2321 pKa = 3.45LTGVNLASHH2330 pKa = 6.36YY2331 pKa = 10.76GVTPGAGGIIGGADD2345 pKa = 3.06TATIINGMLNDD2356 pKa = 4.31HH2357 pKa = 6.08SLKK2360 pKa = 10.51VDD2362 pKa = 3.57TVV2364 pKa = 3.29
Molecular weight: 237.4 kDa
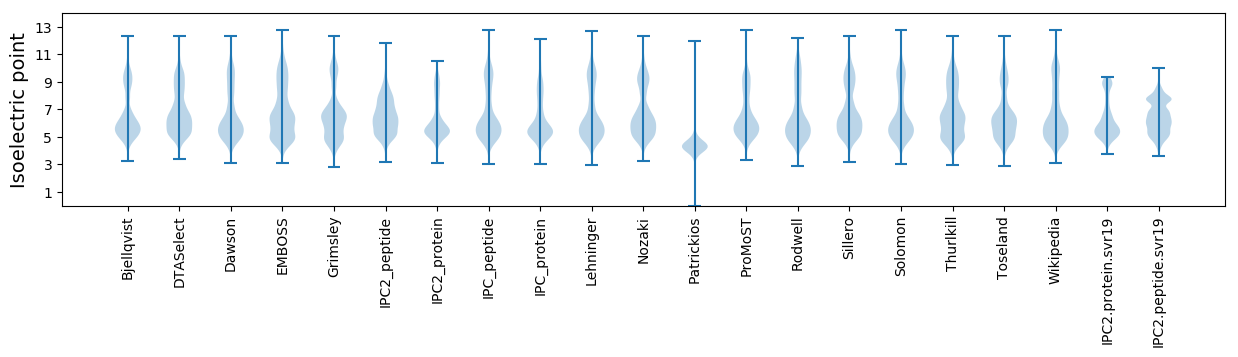
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8Q7Q1|G8Q7Q1_PSEF3 Sulfate-transport transmembrane protein abc transporter OS=Pseudomonas fluorescens (strain F113) OX=1114970 GN=PSF113_3313 PE=3 SV=1
MM1 pKa = 7.12TRR3 pKa = 11.84FFRR6 pKa = 11.84PHH8 pKa = 6.65ALFSHH13 pKa = 6.55ACVHH17 pKa = 5.66GWVLIGLLHH26 pKa = 6.47SASAMAQPDD35 pKa = 3.66VLQDD39 pKa = 3.62KK40 pKa = 9.81VRR42 pKa = 11.84HH43 pKa = 6.31LIAQRR48 pKa = 11.84LPPRR52 pKa = 11.84SQQLQIDD59 pKa = 4.12IGQPAARR66 pKa = 11.84ISACEE71 pKa = 3.84SPLPYY76 pKa = 10.05LVHH79 pKa = 7.09PEE81 pKa = 3.75QNVFGRR87 pKa = 11.84VAVGVRR93 pKa = 11.84CEE95 pKa = 4.0GDD97 pKa = 3.21EE98 pKa = 4.24GVAGYY103 pKa = 9.6LQVSVRR109 pKa = 11.84AVGDD113 pKa = 3.64YY114 pKa = 10.6VISRR118 pKa = 11.84QRR120 pKa = 11.84IAAGDD125 pKa = 3.82VVKK128 pKa = 11.22ADD130 pKa = 3.31MLEE133 pKa = 4.16IKK135 pKa = 10.37RR136 pKa = 11.84GPLEE140 pKa = 4.23RR141 pKa = 11.84LPKK144 pKa = 10.63GSVLKK149 pKa = 10.5PEE151 pKa = 4.14QVLGRR156 pKa = 11.84QASRR160 pKa = 11.84SFSRR164 pKa = 11.84GAALALNNFRR174 pKa = 11.84EE175 pKa = 4.06RR176 pKa = 11.84WLVQRR181 pKa = 11.84NQRR184 pKa = 11.84VVLQARR190 pKa = 11.84GAGFTIRR197 pKa = 11.84RR198 pKa = 11.84DD199 pKa = 3.46GKK201 pKa = 10.89ALDD204 pKa = 3.93NGSLGSSIRR213 pKa = 11.84VLSTDD218 pKa = 3.1GKK220 pKa = 10.57LLNAQVVGQNEE231 pKa = 4.16LLLRR235 pKa = 11.84YY236 pKa = 9.15
MM1 pKa = 7.12TRR3 pKa = 11.84FFRR6 pKa = 11.84PHH8 pKa = 6.65ALFSHH13 pKa = 6.55ACVHH17 pKa = 5.66GWVLIGLLHH26 pKa = 6.47SASAMAQPDD35 pKa = 3.66VLQDD39 pKa = 3.62KK40 pKa = 9.81VRR42 pKa = 11.84HH43 pKa = 6.31LIAQRR48 pKa = 11.84LPPRR52 pKa = 11.84SQQLQIDD59 pKa = 4.12IGQPAARR66 pKa = 11.84ISACEE71 pKa = 3.84SPLPYY76 pKa = 10.05LVHH79 pKa = 7.09PEE81 pKa = 3.75QNVFGRR87 pKa = 11.84VAVGVRR93 pKa = 11.84CEE95 pKa = 4.0GDD97 pKa = 3.21EE98 pKa = 4.24GVAGYY103 pKa = 9.6LQVSVRR109 pKa = 11.84AVGDD113 pKa = 3.64YY114 pKa = 10.6VISRR118 pKa = 11.84QRR120 pKa = 11.84IAAGDD125 pKa = 3.82VVKK128 pKa = 11.22ADD130 pKa = 3.31MLEE133 pKa = 4.16IKK135 pKa = 10.37RR136 pKa = 11.84GPLEE140 pKa = 4.23RR141 pKa = 11.84LPKK144 pKa = 10.63GSVLKK149 pKa = 10.5PEE151 pKa = 4.14QVLGRR156 pKa = 11.84QASRR160 pKa = 11.84SFSRR164 pKa = 11.84GAALALNNFRR174 pKa = 11.84EE175 pKa = 4.06RR176 pKa = 11.84WLVQRR181 pKa = 11.84NQRR184 pKa = 11.84VVLQARR190 pKa = 11.84GAGFTIRR197 pKa = 11.84RR198 pKa = 11.84DD199 pKa = 3.46GKK201 pKa = 10.89ALDD204 pKa = 3.93NGSLGSSIRR213 pKa = 11.84VLSTDD218 pKa = 3.1GKK220 pKa = 10.57LLNAQVVGQNEE231 pKa = 4.16LLLRR235 pKa = 11.84YY236 pKa = 9.15
Molecular weight: 25.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1970731 |
24 |
5414 |
337.6 |
37.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.834 ± 0.037 | 0.989 ± 0.011 |
5.422 ± 0.022 | 5.609 ± 0.029 |
3.673 ± 0.024 | 7.963 ± 0.032 |
2.252 ± 0.017 | 4.837 ± 0.023 |
3.416 ± 0.027 | 11.718 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.016 | 3.102 ± 0.027 |
4.882 ± 0.024 | 4.605 ± 0.025 |
6.31 ± 0.031 | 5.879 ± 0.025 |
5.057 ± 0.049 | 7.167 ± 0.023 |
1.43 ± 0.013 | 2.53 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
