
Methylomusa anaerophila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Methylomusa
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
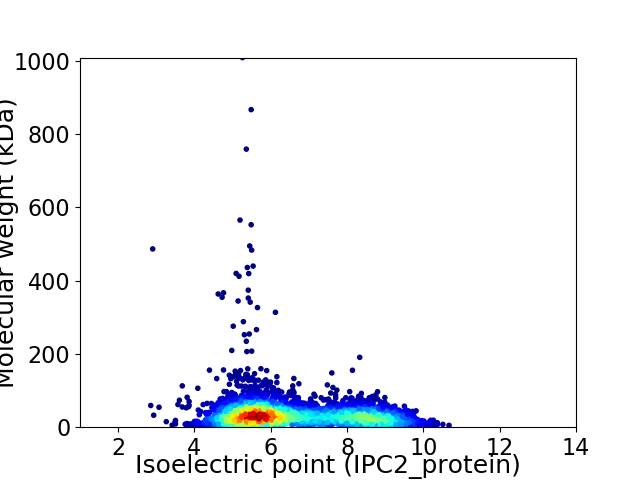
Virtual 2D-PAGE plot for 4152 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A348AKX5|A0A348AKX5_9FIRM Uncharacterized protein OS=Methylomusa anaerophila OX=1930071 GN=MAMMFC1_02407 PE=4 SV=1
MM1 pKa = 7.47ALSTTTLTVAEE12 pKa = 5.21VITKK16 pKa = 9.9AASTDD21 pKa = 2.79SWVIYY26 pKa = 10.63GNTGGEE32 pKa = 4.4NITGSAFADD41 pKa = 3.64TLYY44 pKa = 11.4GLAGADD50 pKa = 3.43TLVGGNGDD58 pKa = 3.93DD59 pKa = 5.07LIVGGTGNDD68 pKa = 3.74TLTGGADD75 pKa = 3.76DD76 pKa = 3.66DD77 pKa = 4.3TFRR80 pKa = 11.84YY81 pKa = 10.22SYY83 pKa = 11.71GDD85 pKa = 3.77GNDD88 pKa = 3.89SIADD92 pKa = 3.67FGVATDD98 pKa = 3.48VDD100 pKa = 3.69ILQFTNIALSNISFNALTAGAAQNITMTGGASIRR134 pKa = 11.84ITNDD138 pKa = 2.21ATANNGNMKK147 pKa = 10.48VITSDD152 pKa = 3.25KK153 pKa = 10.24TFKK156 pKa = 10.88LYY158 pKa = 10.79LDD160 pKa = 3.86NDD162 pKa = 4.21GAPGVVGGSSLADD175 pKa = 3.61FVLGGIGGGSTLIGGTAGADD195 pKa = 3.54TLQGQAGSAGDD206 pKa = 3.7VYY208 pKa = 11.0VYY210 pKa = 10.6RR211 pKa = 11.84SDD213 pKa = 3.66LAKK216 pKa = 10.97VNALGANDD224 pKa = 4.0TLSAAVLSSDD234 pKa = 3.04GTTANNTAVEE244 pKa = 4.11INLYY248 pKa = 7.09DD249 pKa = 3.51TKK251 pKa = 10.63YY252 pKa = 8.68TGVEE256 pKa = 3.99NVVGGGGRR264 pKa = 11.84DD265 pKa = 3.55TLRR268 pKa = 11.84GSSLAEE274 pKa = 3.88TLDD277 pKa = 3.74GGNGGDD283 pKa = 4.22ILWGGAGNDD292 pKa = 3.97SLTGGAGNDD301 pKa = 3.87TYY303 pKa = 11.24WFGTGDD309 pKa = 3.76GLDD312 pKa = 3.84SIVAIGGGGADD323 pKa = 3.78GATDD327 pKa = 3.54ADD329 pKa = 3.98VYY331 pKa = 11.52NFYY334 pKa = 11.2DD335 pKa = 3.52STFSDD340 pKa = 4.6LSFAYY345 pKa = 10.33NGADD349 pKa = 2.98LVVTVGPVTNYY360 pKa = 10.19DD361 pKa = 3.44ALNIQAYY368 pKa = 10.32NNAATNGAKK377 pKa = 9.86IFQTSDD383 pKa = 2.76LTFRR387 pKa = 11.84LLASSAGPIPTGTTAVEE404 pKa = 4.0YY405 pKa = 11.2VRR407 pKa = 11.84DD408 pKa = 3.91FVGAGGVPLLDD419 pKa = 4.18GGGGADD425 pKa = 3.26TVVGGDD431 pKa = 3.3AGNTFAYY438 pKa = 10.44YY439 pKa = 10.97ADD441 pKa = 3.39TARR444 pKa = 11.84YY445 pKa = 8.96IGGSSTLDD453 pKa = 3.16VLTAATSADD462 pKa = 3.12GVEE465 pKa = 3.93INLYY469 pKa = 6.82DD470 pKa = 3.61TKK472 pKa = 11.28YY473 pKa = 10.89SGIEE477 pKa = 3.87KK478 pKa = 8.83VTGSTHH484 pKa = 7.24ADD486 pKa = 3.52TLRR489 pKa = 11.84GTSLADD495 pKa = 3.22SLTGGGGADD504 pKa = 3.99NLWGGAGNDD513 pKa = 4.03TLTGTAGASDD523 pKa = 4.41TYY525 pKa = 11.08WFGTTDD531 pKa = 3.5GNDD534 pKa = 3.47TITAAGADD542 pKa = 3.29ASDD545 pKa = 4.16YY546 pKa = 11.17IFLYY550 pKa = 10.65DD551 pKa = 4.15VSDD554 pKa = 3.47AHH556 pKa = 6.96AQITSMGIEE565 pKa = 4.13GADD568 pKa = 3.28LVLHH572 pKa = 5.86FTGGAALTIAGWTPGGSANRR592 pKa = 11.84FRR594 pKa = 11.84FEE596 pKa = 4.17GNEE599 pKa = 4.01FTWQITDD606 pKa = 3.51PTAANAWARR615 pKa = 11.84VV616 pKa = 3.5
MM1 pKa = 7.47ALSTTTLTVAEE12 pKa = 5.21VITKK16 pKa = 9.9AASTDD21 pKa = 2.79SWVIYY26 pKa = 10.63GNTGGEE32 pKa = 4.4NITGSAFADD41 pKa = 3.64TLYY44 pKa = 11.4GLAGADD50 pKa = 3.43TLVGGNGDD58 pKa = 3.93DD59 pKa = 5.07LIVGGTGNDD68 pKa = 3.74TLTGGADD75 pKa = 3.76DD76 pKa = 3.66DD77 pKa = 4.3TFRR80 pKa = 11.84YY81 pKa = 10.22SYY83 pKa = 11.71GDD85 pKa = 3.77GNDD88 pKa = 3.89SIADD92 pKa = 3.67FGVATDD98 pKa = 3.48VDD100 pKa = 3.69ILQFTNIALSNISFNALTAGAAQNITMTGGASIRR134 pKa = 11.84ITNDD138 pKa = 2.21ATANNGNMKK147 pKa = 10.48VITSDD152 pKa = 3.25KK153 pKa = 10.24TFKK156 pKa = 10.88LYY158 pKa = 10.79LDD160 pKa = 3.86NDD162 pKa = 4.21GAPGVVGGSSLADD175 pKa = 3.61FVLGGIGGGSTLIGGTAGADD195 pKa = 3.54TLQGQAGSAGDD206 pKa = 3.7VYY208 pKa = 11.0VYY210 pKa = 10.6RR211 pKa = 11.84SDD213 pKa = 3.66LAKK216 pKa = 10.97VNALGANDD224 pKa = 4.0TLSAAVLSSDD234 pKa = 3.04GTTANNTAVEE244 pKa = 4.11INLYY248 pKa = 7.09DD249 pKa = 3.51TKK251 pKa = 10.63YY252 pKa = 8.68TGVEE256 pKa = 3.99NVVGGGGRR264 pKa = 11.84DD265 pKa = 3.55TLRR268 pKa = 11.84GSSLAEE274 pKa = 3.88TLDD277 pKa = 3.74GGNGGDD283 pKa = 4.22ILWGGAGNDD292 pKa = 3.97SLTGGAGNDD301 pKa = 3.87TYY303 pKa = 11.24WFGTGDD309 pKa = 3.76GLDD312 pKa = 3.84SIVAIGGGGADD323 pKa = 3.78GATDD327 pKa = 3.54ADD329 pKa = 3.98VYY331 pKa = 11.52NFYY334 pKa = 11.2DD335 pKa = 3.52STFSDD340 pKa = 4.6LSFAYY345 pKa = 10.33NGADD349 pKa = 2.98LVVTVGPVTNYY360 pKa = 10.19DD361 pKa = 3.44ALNIQAYY368 pKa = 10.32NNAATNGAKK377 pKa = 9.86IFQTSDD383 pKa = 2.76LTFRR387 pKa = 11.84LLASSAGPIPTGTTAVEE404 pKa = 4.0YY405 pKa = 11.2VRR407 pKa = 11.84DD408 pKa = 3.91FVGAGGVPLLDD419 pKa = 4.18GGGGADD425 pKa = 3.26TVVGGDD431 pKa = 3.3AGNTFAYY438 pKa = 10.44YY439 pKa = 10.97ADD441 pKa = 3.39TARR444 pKa = 11.84YY445 pKa = 8.96IGGSSTLDD453 pKa = 3.16VLTAATSADD462 pKa = 3.12GVEE465 pKa = 3.93INLYY469 pKa = 6.82DD470 pKa = 3.61TKK472 pKa = 11.28YY473 pKa = 10.89SGIEE477 pKa = 3.87KK478 pKa = 8.83VTGSTHH484 pKa = 7.24ADD486 pKa = 3.52TLRR489 pKa = 11.84GTSLADD495 pKa = 3.22SLTGGGGADD504 pKa = 3.99NLWGGAGNDD513 pKa = 4.03TLTGTAGASDD523 pKa = 4.41TYY525 pKa = 11.08WFGTTDD531 pKa = 3.5GNDD534 pKa = 3.47TITAAGADD542 pKa = 3.29ASDD545 pKa = 4.16YY546 pKa = 11.17IFLYY550 pKa = 10.65DD551 pKa = 4.15VSDD554 pKa = 3.47AHH556 pKa = 6.96AQITSMGIEE565 pKa = 4.13GADD568 pKa = 3.28LVLHH572 pKa = 5.86FTGGAALTIAGWTPGGSANRR592 pKa = 11.84FRR594 pKa = 11.84FEE596 pKa = 4.17GNEE599 pKa = 4.01FTWQITDD606 pKa = 3.51PTAANAWARR615 pKa = 11.84VV616 pKa = 3.5
Molecular weight: 61.8 kDa
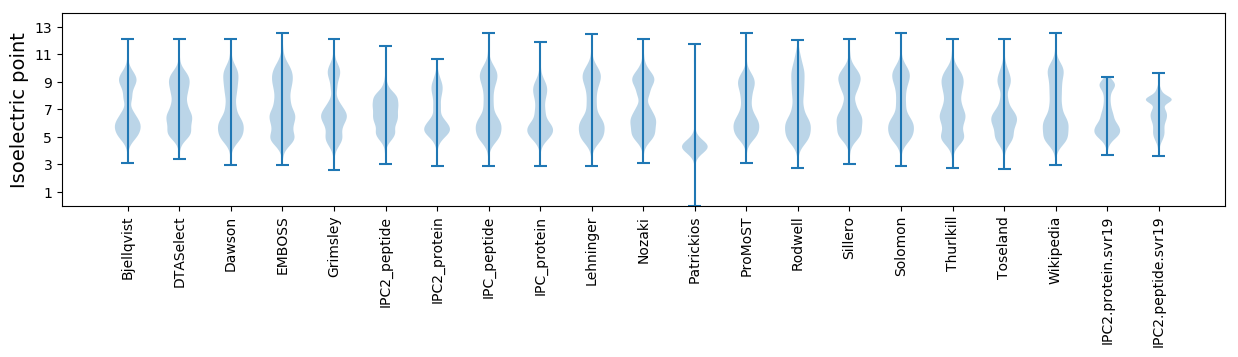
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A348AHW1|A0A348AHW1_9FIRM 34 kDa membrane antigen OS=Methylomusa anaerophila OX=1930071 GN=tpd PE=3 SV=1
MM1 pKa = 6.98SLKK4 pKa = 10.03YY5 pKa = 10.32GKK7 pKa = 9.42YY8 pKa = 8.42VHH10 pKa = 7.09PIVCQTVCQAAGRR23 pKa = 11.84FVILRR28 pKa = 11.84ADD30 pKa = 3.78LEE32 pKa = 4.25NRR34 pKa = 11.84LPAIRR39 pKa = 11.84LLLSTITHH47 pKa = 5.67IRR49 pKa = 11.84VHH51 pKa = 5.72FTCSISVSTHH61 pKa = 5.31APEE64 pKa = 4.62TVNQSQILSQSARR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 3.6
MM1 pKa = 6.98SLKK4 pKa = 10.03YY5 pKa = 10.32GKK7 pKa = 9.42YY8 pKa = 8.42VHH10 pKa = 7.09PIVCQTVCQAAGRR23 pKa = 11.84FVILRR28 pKa = 11.84ADD30 pKa = 3.78LEE32 pKa = 4.25NRR34 pKa = 11.84LPAIRR39 pKa = 11.84LLLSTITHH47 pKa = 5.67IRR49 pKa = 11.84VHH51 pKa = 5.72FTCSISVSTHH61 pKa = 5.31APEE64 pKa = 4.62TVNQSQILSQSARR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 3.6
Molecular weight: 8.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1354701 |
29 |
9197 |
326.3 |
36.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.008 ± 0.052 | 1.194 ± 0.019 |
5.111 ± 0.034 | 6.185 ± 0.045 |
3.873 ± 0.031 | 7.61 ± 0.053 |
1.823 ± 0.018 | 7.339 ± 0.034 |
5.679 ± 0.041 | 9.824 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.02 | 4.331 ± 0.047 |
4.063 ± 0.034 | 3.895 ± 0.037 |
4.868 ± 0.035 | 5.68 ± 0.033 |
5.454 ± 0.042 | 7.128 ± 0.038 |
1.039 ± 0.013 | 3.409 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
