
Epulopiscium sp. SCG-B10WGA-EpuloB
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Epulopiscium; unclassified Epulopiscium
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
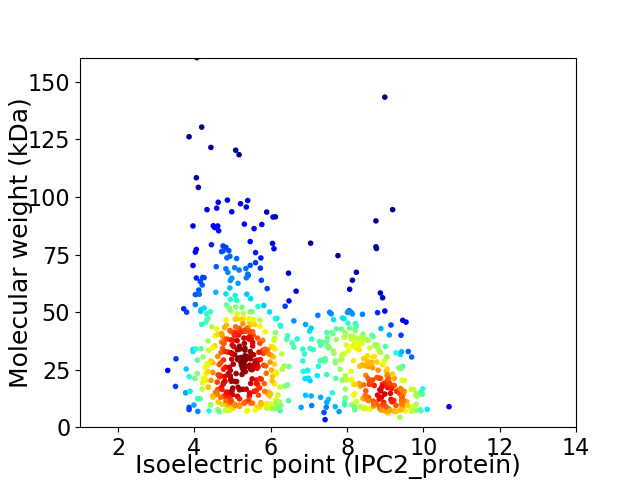
Virtual 2D-PAGE plot for 732 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V2MGA4|A0A1V2MGA4_9FIRM Uncharacterized protein OS=Epulopiscium sp. SCG-B10WGA-EpuloB OX=1712380 GN=AN643_00010 PE=4 SV=1
MM1 pKa = 7.18YY2 pKa = 10.46LSDD5 pKa = 4.22NSIFDD10 pKa = 3.5IYY12 pKa = 11.35NEE14 pKa = 3.97FVGVVDD20 pKa = 5.38EE21 pKa = 6.56DD22 pKa = 3.1IDD24 pKa = 4.26YY25 pKa = 11.7YY26 pKa = 11.35NFNYY30 pKa = 10.62DD31 pKa = 3.74KK32 pKa = 11.44DD33 pKa = 4.49NIDD36 pKa = 4.72SIINNYY42 pKa = 10.59DD43 pKa = 3.51SIFGDD48 pKa = 3.94NILDD52 pKa = 3.61ASTILVADD60 pKa = 3.62EE61 pKa = 4.65TFNSFIDD68 pKa = 4.17FSKK71 pKa = 10.8LSSFEE76 pKa = 3.75FGNIVSQEE84 pKa = 3.83FFEE87 pKa = 4.5EE88 pKa = 4.43HH89 pKa = 6.0LSSYY93 pKa = 10.46YY94 pKa = 10.38VNQVLMYY101 pKa = 8.52MDD103 pKa = 3.86VYY105 pKa = 11.14DD106 pKa = 4.58RR107 pKa = 11.84YY108 pKa = 11.18IMYY111 pKa = 10.6SKK113 pKa = 11.15GDD115 pKa = 3.76DD116 pKa = 3.67YY117 pKa = 11.56FIAYY121 pKa = 9.25YY122 pKa = 10.75NSLEE126 pKa = 4.23SSDD129 pKa = 3.72VRR131 pKa = 11.84YY132 pKa = 7.14TAPGGRR138 pKa = 11.84NTIDD142 pKa = 3.31FNRR145 pKa = 11.84YY146 pKa = 4.78VHH148 pKa = 6.34GFIVKK153 pKa = 8.33FSPDD157 pKa = 3.2YY158 pKa = 10.46GISVEE163 pKa = 4.49SYY165 pKa = 9.79YY166 pKa = 11.28DD167 pKa = 4.34DD168 pKa = 4.36IYY170 pKa = 11.38DD171 pKa = 3.81YY172 pKa = 11.86DD173 pKa = 6.02DD174 pKa = 5.24IIATNDD180 pKa = 3.09YY181 pKa = 10.69RR182 pKa = 11.84VFFVSSAKK190 pKa = 8.9FTYY193 pKa = 10.68DD194 pKa = 4.38DD195 pKa = 4.58ADD197 pKa = 3.8LGLARR202 pKa = 11.84EE203 pKa = 4.33VLYY206 pKa = 11.13GMGDD210 pKa = 3.76LHH212 pKa = 9.11SMIVPPDD219 pKa = 3.34LSFDD223 pKa = 4.01FNQSIEE229 pKa = 4.1NLVVDD234 pKa = 6.05LEE236 pKa = 4.33LTDD239 pKa = 5.89DD240 pKa = 5.54DD241 pKa = 5.36FLEE244 pKa = 4.75LDD246 pKa = 3.8SVLDD250 pKa = 3.46VDD252 pKa = 6.28DD253 pKa = 5.58IISVLRR259 pKa = 11.84DD260 pKa = 3.1KK261 pKa = 11.4DD262 pKa = 3.69LDD264 pKa = 3.79SDD266 pKa = 4.09AGAGMDD272 pKa = 4.05DD273 pKa = 3.87VPVGFFDD280 pKa = 6.73DD281 pKa = 3.46ILSFFDD287 pKa = 4.67NILSSFDD294 pKa = 3.34KK295 pKa = 11.25VLGFFDD301 pKa = 6.57DD302 pKa = 3.87ILSSFDD308 pKa = 3.3KK309 pKa = 11.27VLGFFDD315 pKa = 4.78DD316 pKa = 3.99VFVNLSEE323 pKa = 4.12NLKK326 pKa = 10.84SLFIPDD332 pKa = 3.78FEE334 pKa = 5.63DD335 pKa = 3.07IKK337 pKa = 11.28SRR339 pKa = 11.84FYY341 pKa = 9.11EE342 pKa = 4.26TKK344 pKa = 10.51DD345 pKa = 3.22KK346 pKa = 11.57VSVKK350 pKa = 10.56LGFHH354 pKa = 5.29NWILFEE360 pKa = 5.27DD361 pKa = 4.32ALTLYY366 pKa = 10.51AKK368 pKa = 10.2KK369 pKa = 10.05PKK371 pKa = 10.35FEE373 pKa = 3.91IFIYY377 pKa = 9.58GQAVTLIDD385 pKa = 3.72LSYY388 pKa = 10.76FEE390 pKa = 6.21GYY392 pKa = 8.64MFYY395 pKa = 10.74LQDD398 pKa = 3.71IIKK401 pKa = 10.52GFFSFLLLLYY411 pKa = 9.28NYY413 pKa = 10.78KK414 pKa = 9.61MVLYY418 pKa = 9.99LLRR421 pKa = 11.84SSHH424 pKa = 6.96FIKK427 pKa = 10.66GGKK430 pKa = 8.53
MM1 pKa = 7.18YY2 pKa = 10.46LSDD5 pKa = 4.22NSIFDD10 pKa = 3.5IYY12 pKa = 11.35NEE14 pKa = 3.97FVGVVDD20 pKa = 5.38EE21 pKa = 6.56DD22 pKa = 3.1IDD24 pKa = 4.26YY25 pKa = 11.7YY26 pKa = 11.35NFNYY30 pKa = 10.62DD31 pKa = 3.74KK32 pKa = 11.44DD33 pKa = 4.49NIDD36 pKa = 4.72SIINNYY42 pKa = 10.59DD43 pKa = 3.51SIFGDD48 pKa = 3.94NILDD52 pKa = 3.61ASTILVADD60 pKa = 3.62EE61 pKa = 4.65TFNSFIDD68 pKa = 4.17FSKK71 pKa = 10.8LSSFEE76 pKa = 3.75FGNIVSQEE84 pKa = 3.83FFEE87 pKa = 4.5EE88 pKa = 4.43HH89 pKa = 6.0LSSYY93 pKa = 10.46YY94 pKa = 10.38VNQVLMYY101 pKa = 8.52MDD103 pKa = 3.86VYY105 pKa = 11.14DD106 pKa = 4.58RR107 pKa = 11.84YY108 pKa = 11.18IMYY111 pKa = 10.6SKK113 pKa = 11.15GDD115 pKa = 3.76DD116 pKa = 3.67YY117 pKa = 11.56FIAYY121 pKa = 9.25YY122 pKa = 10.75NSLEE126 pKa = 4.23SSDD129 pKa = 3.72VRR131 pKa = 11.84YY132 pKa = 7.14TAPGGRR138 pKa = 11.84NTIDD142 pKa = 3.31FNRR145 pKa = 11.84YY146 pKa = 4.78VHH148 pKa = 6.34GFIVKK153 pKa = 8.33FSPDD157 pKa = 3.2YY158 pKa = 10.46GISVEE163 pKa = 4.49SYY165 pKa = 9.79YY166 pKa = 11.28DD167 pKa = 4.34DD168 pKa = 4.36IYY170 pKa = 11.38DD171 pKa = 3.81YY172 pKa = 11.86DD173 pKa = 6.02DD174 pKa = 5.24IIATNDD180 pKa = 3.09YY181 pKa = 10.69RR182 pKa = 11.84VFFVSSAKK190 pKa = 8.9FTYY193 pKa = 10.68DD194 pKa = 4.38DD195 pKa = 4.58ADD197 pKa = 3.8LGLARR202 pKa = 11.84EE203 pKa = 4.33VLYY206 pKa = 11.13GMGDD210 pKa = 3.76LHH212 pKa = 9.11SMIVPPDD219 pKa = 3.34LSFDD223 pKa = 4.01FNQSIEE229 pKa = 4.1NLVVDD234 pKa = 6.05LEE236 pKa = 4.33LTDD239 pKa = 5.89DD240 pKa = 5.54DD241 pKa = 5.36FLEE244 pKa = 4.75LDD246 pKa = 3.8SVLDD250 pKa = 3.46VDD252 pKa = 6.28DD253 pKa = 5.58IISVLRR259 pKa = 11.84DD260 pKa = 3.1KK261 pKa = 11.4DD262 pKa = 3.69LDD264 pKa = 3.79SDD266 pKa = 4.09AGAGMDD272 pKa = 4.05DD273 pKa = 3.87VPVGFFDD280 pKa = 6.73DD281 pKa = 3.46ILSFFDD287 pKa = 4.67NILSSFDD294 pKa = 3.34KK295 pKa = 11.25VLGFFDD301 pKa = 6.57DD302 pKa = 3.87ILSSFDD308 pKa = 3.3KK309 pKa = 11.27VLGFFDD315 pKa = 4.78DD316 pKa = 3.99VFVNLSEE323 pKa = 4.12NLKK326 pKa = 10.84SLFIPDD332 pKa = 3.78FEE334 pKa = 5.63DD335 pKa = 3.07IKK337 pKa = 11.28SRR339 pKa = 11.84FYY341 pKa = 9.11EE342 pKa = 4.26TKK344 pKa = 10.51DD345 pKa = 3.22KK346 pKa = 11.57VSVKK350 pKa = 10.56LGFHH354 pKa = 5.29NWILFEE360 pKa = 5.27DD361 pKa = 4.32ALTLYY366 pKa = 10.51AKK368 pKa = 10.2KK369 pKa = 10.05PKK371 pKa = 10.35FEE373 pKa = 3.91IFIYY377 pKa = 9.58GQAVTLIDD385 pKa = 3.72LSYY388 pKa = 10.76FEE390 pKa = 6.21GYY392 pKa = 8.64MFYY395 pKa = 10.74LQDD398 pKa = 3.71IIKK401 pKa = 10.52GFFSFLLLLYY411 pKa = 9.28NYY413 pKa = 10.78KK414 pKa = 9.61MVLYY418 pKa = 9.99LLRR421 pKa = 11.84SSHH424 pKa = 6.96FIKK427 pKa = 10.66GGKK430 pKa = 8.53
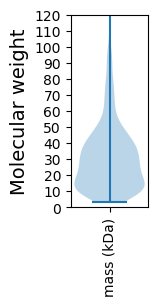
Molecular weight: 49.97 kDa
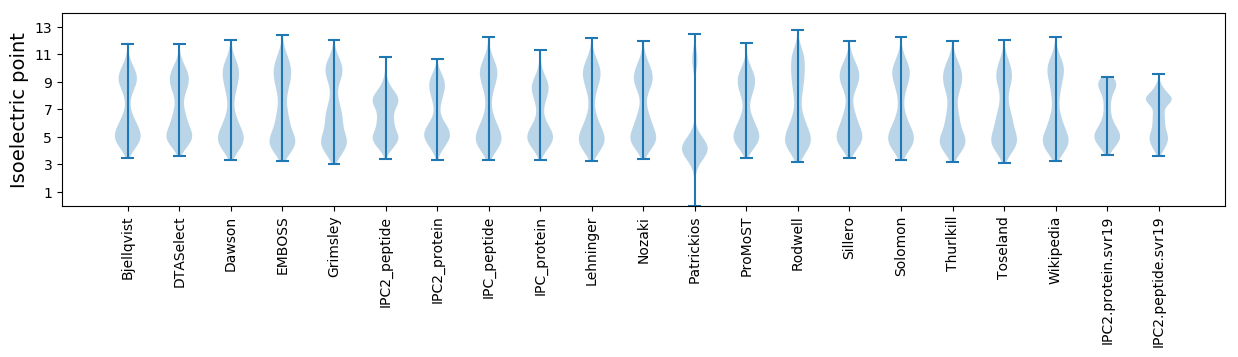
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V2MFA3|A0A1V2MFA3_9FIRM PRC domain-containing protein OS=Epulopiscium sp. SCG-B10WGA-EpuloB OX=1712380 GN=AN643_00935 PE=4 SV=1
MM1 pKa = 7.3TEE3 pKa = 3.95KK4 pKa = 10.52EE5 pKa = 3.83YY6 pKa = 11.26DD7 pKa = 3.37LSIFKK12 pKa = 10.42IKK14 pKa = 10.54EE15 pKa = 3.99FVHH18 pKa = 6.7LPLALEE24 pKa = 4.52LSHH27 pKa = 6.79GLLLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84CHH36 pKa = 5.78EE37 pKa = 3.86VTEE40 pKa = 4.69GVGGRR45 pKa = 11.84WQRR48 pKa = 11.84YY49 pKa = 4.54STLPLRR55 pKa = 11.84GVPHH59 pKa = 5.65QRR61 pKa = 11.84GGLYY65 pKa = 10.29GWSKK69 pKa = 8.88RR70 pKa = 11.84AKK72 pKa = 9.98RR73 pKa = 11.84RR74 pKa = 11.84TT75 pKa = 3.39
MM1 pKa = 7.3TEE3 pKa = 3.95KK4 pKa = 10.52EE5 pKa = 3.83YY6 pKa = 11.26DD7 pKa = 3.37LSIFKK12 pKa = 10.42IKK14 pKa = 10.54EE15 pKa = 3.99FVHH18 pKa = 6.7LPLALEE24 pKa = 4.52LSHH27 pKa = 6.79GLLLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84CHH36 pKa = 5.78EE37 pKa = 3.86VTEE40 pKa = 4.69GVGGRR45 pKa = 11.84WQRR48 pKa = 11.84YY49 pKa = 4.54STLPLRR55 pKa = 11.84GVPHH59 pKa = 5.65QRR61 pKa = 11.84GGLYY65 pKa = 10.29GWSKK69 pKa = 8.88RR70 pKa = 11.84AKK72 pKa = 9.98RR73 pKa = 11.84RR74 pKa = 11.84TT75 pKa = 3.39
Molecular weight: 8.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
208488 |
30 |
1434 |
284.8 |
32.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.126 ± 0.1 | 1.053 ± 0.041 |
5.852 ± 0.092 | 7.334 ± 0.114 |
4.208 ± 0.074 | 6.098 ± 0.122 |
1.612 ± 0.036 | 9.503 ± 0.117 |
7.924 ± 0.139 | 9.068 ± 0.1 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.045 | 6.013 ± 0.084 |
3.233 ± 0.078 | 3.188 ± 0.065 |
3.389 ± 0.07 | 6.284 ± 0.084 |
5.649 ± 0.091 | 5.981 ± 0.073 |
0.73 ± 0.03 | 4.114 ± 0.073 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
