
Erysiphe pulchra
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Erysiphales; Erysiphaceae; Erysiphe
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
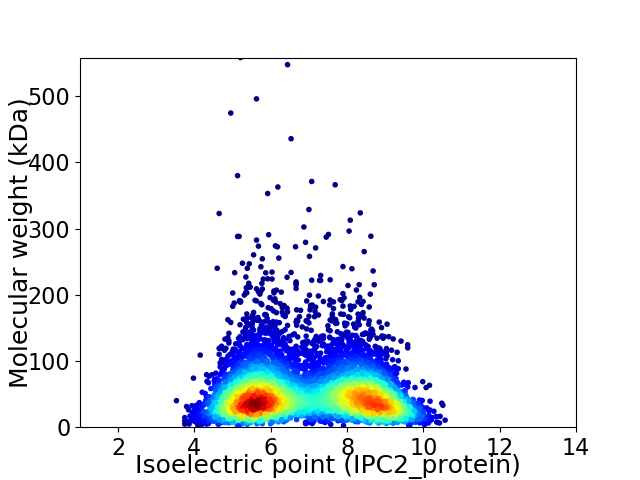
Virtual 2D-PAGE plot for 6853 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
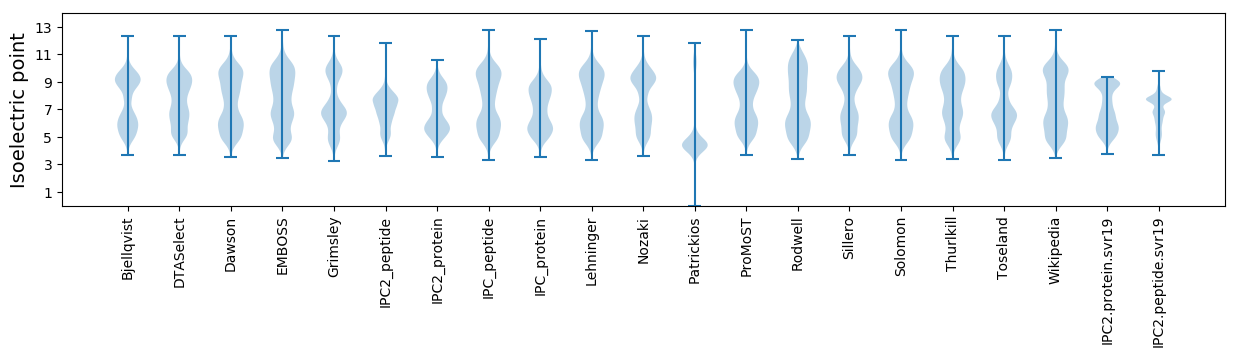
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S4PTU3|A0A2S4PTU3_9PEZI SURF6-domain-containing protein (Fragment) OS=Erysiphe pulchra OX=225359 GN=EPUL_003951 PE=3 SV=1
MM1 pKa = 7.42TGATSEE7 pKa = 3.93AGYY10 pKa = 10.93DD11 pKa = 3.45VVVDD15 pKa = 3.65VDD17 pKa = 3.45EE18 pKa = 6.42DD19 pKa = 4.15GDD21 pKa = 4.32LGHH24 pKa = 7.38TDD26 pKa = 3.4LQEE29 pKa = 4.41DD30 pKa = 4.7LEE32 pKa = 4.49FHH34 pKa = 7.04SSNFNNDD41 pKa = 2.2ASTASNSRR49 pKa = 11.84KK50 pKa = 10.23GPGAGTGLPIPATANTGSPKK70 pKa = 10.52YY71 pKa = 9.24MIWSLPFYY79 pKa = 10.89AQFFDD84 pKa = 3.45VDD86 pKa = 3.79TAAVLSRR93 pKa = 11.84CWATIYY99 pKa = 10.37PRR101 pKa = 11.84ANFIEE106 pKa = 4.25VLGGNPDD113 pKa = 3.24LYY115 pKa = 11.23GPLWIATTVVFILFLGGTINQYY137 pKa = 10.95LSEE140 pKa = 4.4TNGTQFVYY148 pKa = 10.83DD149 pKa = 4.04FTLLSGG155 pKa = 3.76
MM1 pKa = 7.42TGATSEE7 pKa = 3.93AGYY10 pKa = 10.93DD11 pKa = 3.45VVVDD15 pKa = 3.65VDD17 pKa = 3.45EE18 pKa = 6.42DD19 pKa = 4.15GDD21 pKa = 4.32LGHH24 pKa = 7.38TDD26 pKa = 3.4LQEE29 pKa = 4.41DD30 pKa = 4.7LEE32 pKa = 4.49FHH34 pKa = 7.04SSNFNNDD41 pKa = 2.2ASTASNSRR49 pKa = 11.84KK50 pKa = 10.23GPGAGTGLPIPATANTGSPKK70 pKa = 10.52YY71 pKa = 9.24MIWSLPFYY79 pKa = 10.89AQFFDD84 pKa = 3.45VDD86 pKa = 3.79TAAVLSRR93 pKa = 11.84CWATIYY99 pKa = 10.37PRR101 pKa = 11.84ANFIEE106 pKa = 4.25VLGGNPDD113 pKa = 3.24LYY115 pKa = 11.23GPLWIATTVVFILFLGGTINQYY137 pKa = 10.95LSEE140 pKa = 4.4TNGTQFVYY148 pKa = 10.83DD149 pKa = 4.04FTLLSGG155 pKa = 3.76
Molecular weight: 16.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S4PIJ3|A0A2S4PIJ3_9PEZI Uncharacterized protein (Fragment) OS=Erysiphe pulchra OX=225359 GN=EPUL_006201 PE=4 SV=1
MM1 pKa = 7.73ALTFPVPSTQQILQTCLSAAKK22 pKa = 9.73RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84SSSSEE30 pKa = 3.61KK31 pKa = 10.73APTSRR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.89ARR41 pKa = 11.84SSRR44 pKa = 11.84TSTHH48 pKa = 6.62ASPSRR53 pKa = 11.84QLSRR57 pKa = 11.84ALSVPTAVIAARR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84KK72 pKa = 8.33WQTNDD77 pKa = 2.96YY78 pKa = 9.65QRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84DD83 pKa = 3.26SHH85 pKa = 5.49EE86 pKa = 4.16APRR89 pKa = 11.84HH90 pKa = 4.44CAPRR94 pKa = 11.84RR95 pKa = 11.84TSTRR99 pKa = 11.84RR100 pKa = 11.84HH101 pKa = 5.65CEE103 pKa = 3.14ITGRR107 pKa = 11.84RR108 pKa = 11.84GGDD111 pKa = 3.0GTTSVGGSGGRR122 pKa = 11.84DD123 pKa = 3.3PEE125 pKa = 4.57GGQGVRR131 pKa = 11.84RR132 pKa = 11.84GGRR135 pKa = 11.84PEE137 pKa = 3.87KK138 pKa = 10.44SEE140 pKa = 4.37HH141 pKa = 6.16NGGEE145 pKa = 3.87QGQGDD150 pKa = 3.95RR151 pKa = 11.84GQHH154 pKa = 5.17EE155 pKa = 4.43RR156 pKa = 11.84EE157 pKa = 4.03QPEE160 pKa = 4.02GNAAEE165 pKa = 4.47TRR167 pKa = 11.84RR168 pKa = 11.84HH169 pKa = 4.19GHH171 pKa = 5.19VLEE174 pKa = 4.5TDD176 pKa = 3.85PPQLRR181 pKa = 11.84LFHH184 pKa = 7.17EE185 pKa = 5.08DD186 pKa = 2.96GGGCRR191 pKa = 11.84AHH193 pKa = 6.8SGPEE197 pKa = 3.73RR198 pKa = 11.84PEE200 pKa = 3.75QQAYY204 pKa = 7.17WCQEE208 pKa = 3.73GYY210 pKa = 9.91RR211 pKa = 11.84WCAPGLALRR220 pKa = 11.84QRR222 pKa = 11.84CGFQEE227 pKa = 4.98DD228 pKa = 3.46ILVRR232 pKa = 11.84RR233 pKa = 11.84FRR235 pKa = 11.84AAAQVFQEE243 pKa = 4.83PEE245 pKa = 3.8DD246 pKa = 3.93CLLEE250 pKa = 4.89RR251 pKa = 11.84GAGTEE256 pKa = 4.2GG257 pKa = 3.21
MM1 pKa = 7.73ALTFPVPSTQQILQTCLSAAKK22 pKa = 9.73RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84SSSSEE30 pKa = 3.61KK31 pKa = 10.73APTSRR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.89ARR41 pKa = 11.84SSRR44 pKa = 11.84TSTHH48 pKa = 6.62ASPSRR53 pKa = 11.84QLSRR57 pKa = 11.84ALSVPTAVIAARR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84KK72 pKa = 8.33WQTNDD77 pKa = 2.96YY78 pKa = 9.65QRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84DD83 pKa = 3.26SHH85 pKa = 5.49EE86 pKa = 4.16APRR89 pKa = 11.84HH90 pKa = 4.44CAPRR94 pKa = 11.84RR95 pKa = 11.84TSTRR99 pKa = 11.84RR100 pKa = 11.84HH101 pKa = 5.65CEE103 pKa = 3.14ITGRR107 pKa = 11.84RR108 pKa = 11.84GGDD111 pKa = 3.0GTTSVGGSGGRR122 pKa = 11.84DD123 pKa = 3.3PEE125 pKa = 4.57GGQGVRR131 pKa = 11.84RR132 pKa = 11.84GGRR135 pKa = 11.84PEE137 pKa = 3.87KK138 pKa = 10.44SEE140 pKa = 4.37HH141 pKa = 6.16NGGEE145 pKa = 3.87QGQGDD150 pKa = 3.95RR151 pKa = 11.84GQHH154 pKa = 5.17EE155 pKa = 4.43RR156 pKa = 11.84EE157 pKa = 4.03QPEE160 pKa = 4.02GNAAEE165 pKa = 4.47TRR167 pKa = 11.84RR168 pKa = 11.84HH169 pKa = 4.19GHH171 pKa = 5.19VLEE174 pKa = 4.5TDD176 pKa = 3.85PPQLRR181 pKa = 11.84LFHH184 pKa = 7.17EE185 pKa = 5.08DD186 pKa = 2.96GGGCRR191 pKa = 11.84AHH193 pKa = 6.8SGPEE197 pKa = 3.73RR198 pKa = 11.84PEE200 pKa = 3.75QQAYY204 pKa = 7.17WCQEE208 pKa = 3.73GYY210 pKa = 9.91RR211 pKa = 11.84WCAPGLALRR220 pKa = 11.84QRR222 pKa = 11.84CGFQEE227 pKa = 4.98DD228 pKa = 3.46ILVRR232 pKa = 11.84RR233 pKa = 11.84FRR235 pKa = 11.84AAAQVFQEE243 pKa = 4.83PEE245 pKa = 3.8DD246 pKa = 3.93CLLEE250 pKa = 4.89RR251 pKa = 11.84GAGTEE256 pKa = 4.2GG257 pKa = 3.21
Molecular weight: 28.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3364779 |
18 |
4885 |
491.0 |
55.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.446 ± 0.025 | 1.332 ± 0.011 |
5.45 ± 0.018 | 6.545 ± 0.031 |
3.884 ± 0.017 | 5.505 ± 0.024 |
2.257 ± 0.012 | 6.542 ± 0.02 |
6.455 ± 0.028 | 9.17 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.935 ± 0.01 | 5.165 ± 0.02 |
5.098 ± 0.024 | 4.101 ± 0.021 |
5.734 ± 0.022 | 9.192 ± 0.036 |
5.777 ± 0.016 | 5.34 ± 0.017 |
1.22 ± 0.011 | 2.853 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
