
Oleiphilus sp. HI0079
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oleiphilaceae; Oleiphilus; unclassified Oleiphilus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
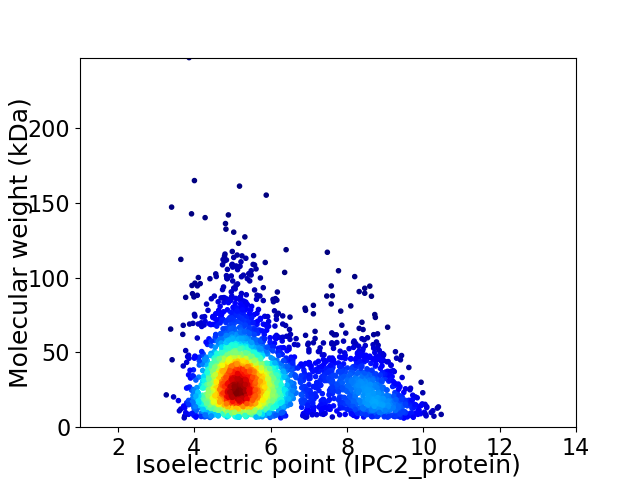
Virtual 2D-PAGE plot for 3311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A657BAF4|A0A657BAF4_9GAMM Uncharacterized protein OS=Oleiphilus sp. HI0079 OX=1822254 GN=A3750_11315 PE=4 SV=1
MM1 pKa = 7.11YY2 pKa = 10.11LKK4 pKa = 9.87PQPGWSKK11 pKa = 11.02LSLAVAVAGALSLGGCKK28 pKa = 7.77VTIKK32 pKa = 10.61ADD34 pKa = 3.4EE35 pKa = 4.9GEE37 pKa = 4.17VSDD40 pKa = 5.09IVDD43 pKa = 3.8SVTDD47 pKa = 3.61NAVCRR52 pKa = 11.84DD53 pKa = 3.5DD54 pKa = 5.65DD55 pKa = 4.55GNKK58 pKa = 9.35VEE60 pKa = 5.3CGNSDD65 pKa = 3.5GGSATVLSASGYY77 pKa = 8.61EE78 pKa = 3.98IPEE81 pKa = 4.04DD82 pKa = 4.54AIFVAADD89 pKa = 3.36ARR91 pKa = 11.84DD92 pKa = 3.69GDD94 pKa = 5.26DD95 pKa = 3.28ITDD98 pKa = 5.36AINLALFEE106 pKa = 4.48VPAGSTIGLPKK117 pKa = 10.21GTFKK121 pKa = 10.93VSNSIIVYY129 pKa = 8.77NANDD133 pKa = 3.68ITLMGYY139 pKa = 10.07GINDD143 pKa = 3.64TVLDD147 pKa = 4.09FDD149 pKa = 5.29GSSGDD154 pKa = 3.34DD155 pKa = 3.19GFRR158 pKa = 11.84FEE160 pKa = 5.01GGSDD164 pKa = 3.52LMVRR168 pKa = 11.84DD169 pKa = 4.02LGVYY173 pKa = 7.63DD174 pKa = 3.83TNKK177 pKa = 10.65NGIKK181 pKa = 9.29TVGSNGVYY189 pKa = 9.92FSHH192 pKa = 6.84VAAVWTVGRR201 pKa = 11.84LSDD204 pKa = 3.82EE205 pKa = 4.21TPKK208 pKa = 10.87RR209 pKa = 11.84LLADD213 pKa = 3.39SPGAYY218 pKa = 9.75GLYY221 pKa = 9.73PVNSKK226 pKa = 11.15NIVIEE231 pKa = 4.21DD232 pKa = 3.83TFSCGSRR239 pKa = 11.84DD240 pKa = 3.17AGIYY244 pKa = 9.57VGQSEE249 pKa = 4.65DD250 pKa = 3.5VVVRR254 pKa = 11.84NNLVEE259 pKa = 4.17YY260 pKa = 9.7NVAGIEE266 pKa = 4.46IEE268 pKa = 4.13NTEE271 pKa = 4.15NADD274 pKa = 3.45VYY276 pKa = 11.8NNDD279 pKa = 3.51VYY281 pKa = 11.72DD282 pKa = 4.07NTAGWSPVTRR292 pKa = 11.84NINIHH297 pKa = 6.01NNSLSQNSTDD307 pKa = 3.57PGGKK311 pKa = 9.43LLEE314 pKa = 4.35EE315 pKa = 4.71AGVIAGYY322 pKa = 8.97EE323 pKa = 4.04AYY325 pKa = 10.27YY326 pKa = 10.68GAGSFPDD333 pKa = 3.38ILYY336 pKa = 10.88GGIGEE341 pKa = 4.73LLANAGEE348 pKa = 4.15LAPLGIAPYY357 pKa = 10.42AADD360 pKa = 4.25GSDD363 pKa = 3.87NVCEE367 pKa = 4.27TNNFEE372 pKa = 4.36AQQGAADD379 pKa = 3.98EE380 pKa = 4.67ASVGLVYY387 pKa = 9.32GTNPMDD393 pKa = 4.46PNNWVWDD400 pKa = 4.09GTPGASNILGPQATLSRR417 pKa = 11.84EE418 pKa = 4.06LLSANANMNCSLPRR432 pKa = 11.84LAGSSVVFRR441 pKa = 11.84GEE443 pKa = 3.92EE444 pKa = 4.05FGCNSDD450 pKa = 5.6DD451 pKa = 4.08LDD453 pKa = 4.54EE454 pKa = 4.59YY455 pKa = 11.62ACSLL459 pKa = 3.82
MM1 pKa = 7.11YY2 pKa = 10.11LKK4 pKa = 9.87PQPGWSKK11 pKa = 11.02LSLAVAVAGALSLGGCKK28 pKa = 7.77VTIKK32 pKa = 10.61ADD34 pKa = 3.4EE35 pKa = 4.9GEE37 pKa = 4.17VSDD40 pKa = 5.09IVDD43 pKa = 3.8SVTDD47 pKa = 3.61NAVCRR52 pKa = 11.84DD53 pKa = 3.5DD54 pKa = 5.65DD55 pKa = 4.55GNKK58 pKa = 9.35VEE60 pKa = 5.3CGNSDD65 pKa = 3.5GGSATVLSASGYY77 pKa = 8.61EE78 pKa = 3.98IPEE81 pKa = 4.04DD82 pKa = 4.54AIFVAADD89 pKa = 3.36ARR91 pKa = 11.84DD92 pKa = 3.69GDD94 pKa = 5.26DD95 pKa = 3.28ITDD98 pKa = 5.36AINLALFEE106 pKa = 4.48VPAGSTIGLPKK117 pKa = 10.21GTFKK121 pKa = 10.93VSNSIIVYY129 pKa = 8.77NANDD133 pKa = 3.68ITLMGYY139 pKa = 10.07GINDD143 pKa = 3.64TVLDD147 pKa = 4.09FDD149 pKa = 5.29GSSGDD154 pKa = 3.34DD155 pKa = 3.19GFRR158 pKa = 11.84FEE160 pKa = 5.01GGSDD164 pKa = 3.52LMVRR168 pKa = 11.84DD169 pKa = 4.02LGVYY173 pKa = 7.63DD174 pKa = 3.83TNKK177 pKa = 10.65NGIKK181 pKa = 9.29TVGSNGVYY189 pKa = 9.92FSHH192 pKa = 6.84VAAVWTVGRR201 pKa = 11.84LSDD204 pKa = 3.82EE205 pKa = 4.21TPKK208 pKa = 10.87RR209 pKa = 11.84LLADD213 pKa = 3.39SPGAYY218 pKa = 9.75GLYY221 pKa = 9.73PVNSKK226 pKa = 11.15NIVIEE231 pKa = 4.21DD232 pKa = 3.83TFSCGSRR239 pKa = 11.84DD240 pKa = 3.17AGIYY244 pKa = 9.57VGQSEE249 pKa = 4.65DD250 pKa = 3.5VVVRR254 pKa = 11.84NNLVEE259 pKa = 4.17YY260 pKa = 9.7NVAGIEE266 pKa = 4.46IEE268 pKa = 4.13NTEE271 pKa = 4.15NADD274 pKa = 3.45VYY276 pKa = 11.8NNDD279 pKa = 3.51VYY281 pKa = 11.72DD282 pKa = 4.07NTAGWSPVTRR292 pKa = 11.84NINIHH297 pKa = 6.01NNSLSQNSTDD307 pKa = 3.57PGGKK311 pKa = 9.43LLEE314 pKa = 4.35EE315 pKa = 4.71AGVIAGYY322 pKa = 8.97EE323 pKa = 4.04AYY325 pKa = 10.27YY326 pKa = 10.68GAGSFPDD333 pKa = 3.38ILYY336 pKa = 10.88GGIGEE341 pKa = 4.73LLANAGEE348 pKa = 4.15LAPLGIAPYY357 pKa = 10.42AADD360 pKa = 4.25GSDD363 pKa = 3.87NVCEE367 pKa = 4.27TNNFEE372 pKa = 4.36AQQGAADD379 pKa = 3.98EE380 pKa = 4.67ASVGLVYY387 pKa = 9.32GTNPMDD393 pKa = 4.46PNNWVWDD400 pKa = 4.09GTPGASNILGPQATLSRR417 pKa = 11.84EE418 pKa = 4.06LLSANANMNCSLPRR432 pKa = 11.84LAGSSVVFRR441 pKa = 11.84GEE443 pKa = 3.92EE444 pKa = 4.05FGCNSDD450 pKa = 5.6DD451 pKa = 4.08LDD453 pKa = 4.54EE454 pKa = 4.59YY455 pKa = 11.62ACSLL459 pKa = 3.82
Molecular weight: 48.09 kDa
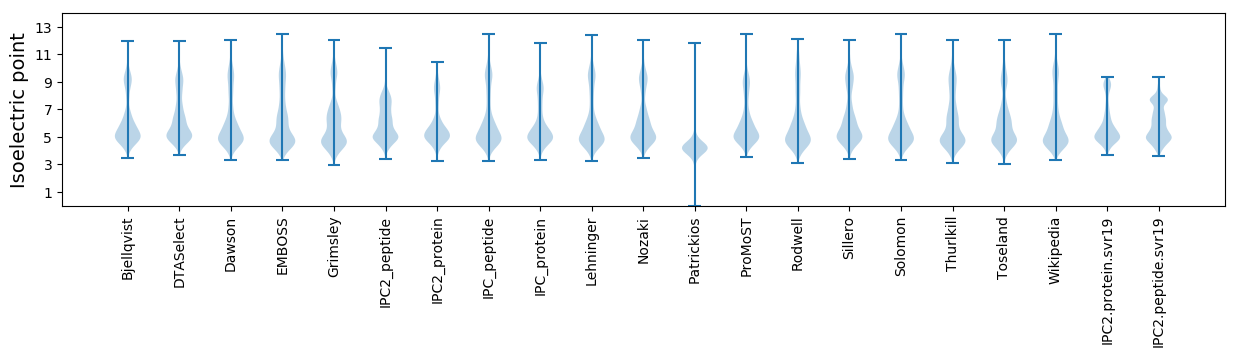
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A657BC37|A0A657BC37_9GAMM Nitrate ABC transporter ATP-binding protein OS=Oleiphilus sp. HI0079 OX=1822254 GN=A3750_09320 PE=4 SV=1
MM1 pKa = 7.58GIEE4 pKa = 3.84TAKK7 pKa = 10.58AAQHH11 pKa = 5.73PKK13 pKa = 8.33MRR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 7.93DD18 pKa = 3.63TRR20 pKa = 11.84PSNVFHH26 pKa = 6.78LSSRR30 pKa = 11.84SMLRR34 pKa = 11.84LHH36 pKa = 7.06GMNLNFGEE44 pKa = 4.41LPRR47 pKa = 11.84FNLQIRR53 pKa = 11.84VFNFTISNFLLDD65 pKa = 3.66WRR67 pKa = 11.84AHH69 pKa = 5.68EE70 pKa = 4.33SSEE73 pKa = 4.14
MM1 pKa = 7.58GIEE4 pKa = 3.84TAKK7 pKa = 10.58AAQHH11 pKa = 5.73PKK13 pKa = 8.33MRR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 7.93DD18 pKa = 3.63TRR20 pKa = 11.84PSNVFHH26 pKa = 6.78LSSRR30 pKa = 11.84SMLRR34 pKa = 11.84LHH36 pKa = 7.06GMNLNFGEE44 pKa = 4.41LPRR47 pKa = 11.84FNLQIRR53 pKa = 11.84VFNFTISNFLLDD65 pKa = 3.66WRR67 pKa = 11.84AHH69 pKa = 5.68EE70 pKa = 4.33SSEE73 pKa = 4.14
Molecular weight: 8.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015081 |
51 |
2284 |
306.6 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.092 ± 0.04 | 1.016 ± 0.012 |
5.826 ± 0.038 | 6.808 ± 0.042 |
4.153 ± 0.024 | 6.955 ± 0.044 |
2.161 ± 0.022 | 6.006 ± 0.033 |
5.041 ± 0.038 | 10.404 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.021 | 3.859 ± 0.029 |
3.916 ± 0.026 | 4.093 ± 0.028 |
5.064 ± 0.033 | 7.108 ± 0.036 |
4.971 ± 0.037 | 7.027 ± 0.039 |
1.121 ± 0.015 | 2.909 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
