
Porcine transmissible gastroenteritis coronavirus (strain Purdue) (TGEV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Tegacovirus; Alphacoronavirus 1; Transmissible gastroenteritis virus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
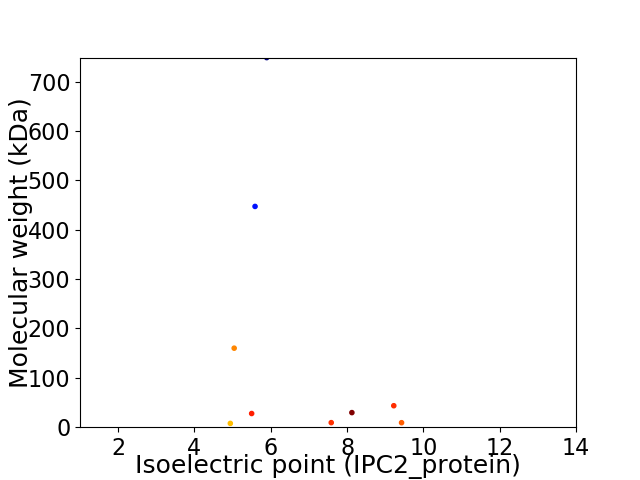
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
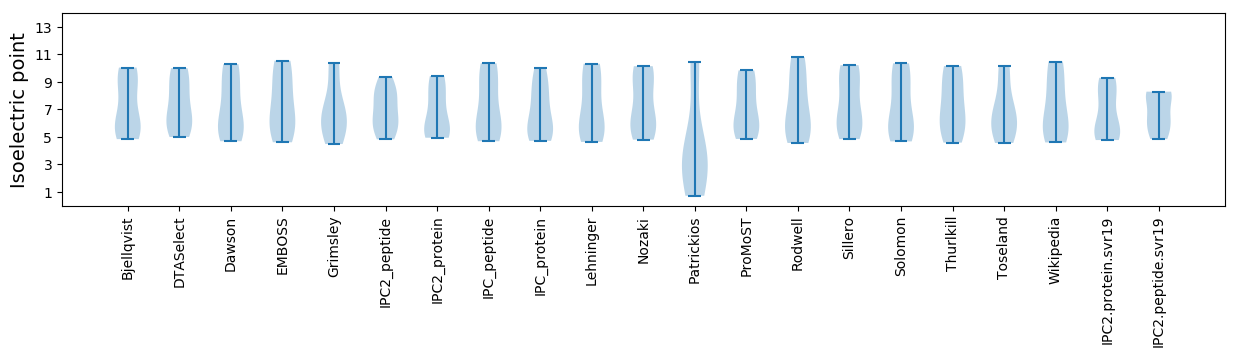
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P09047|NS3B_CVPPU Putative non-structural protein 3b OS=Porcine transmissible gastroenteritis coronavirus (strain Purdue) OX=11151 GN=3b PE=5 SV=2
MM1 pKa = 8.02DD2 pKa = 4.08IVKK5 pKa = 10.35SIYY8 pKa = 9.68TSVDD12 pKa = 2.93AVLDD16 pKa = 4.08EE17 pKa = 4.83LDD19 pKa = 3.58CAYY22 pKa = 10.56FAVTLKK28 pKa = 11.12VEE30 pKa = 4.99FKK32 pKa = 9.86TGKK35 pKa = 10.29LLVCIGFGDD44 pKa = 3.78TLLAAKK50 pKa = 10.2DD51 pKa = 3.25KK52 pKa = 11.08AYY54 pKa = 10.71AKK56 pKa = 10.16LGLSIIEE63 pKa = 4.36EE64 pKa = 4.51VNSHH68 pKa = 5.7IVVV71 pKa = 3.59
MM1 pKa = 8.02DD2 pKa = 4.08IVKK5 pKa = 10.35SIYY8 pKa = 9.68TSVDD12 pKa = 2.93AVLDD16 pKa = 4.08EE17 pKa = 4.83LDD19 pKa = 3.58CAYY22 pKa = 10.56FAVTLKK28 pKa = 11.12VEE30 pKa = 4.99FKK32 pKa = 9.86TGKK35 pKa = 10.29LLVCIGFGDD44 pKa = 3.78TLLAAKK50 pKa = 10.2DD51 pKa = 3.25KK52 pKa = 11.08AYY54 pKa = 10.71AKK56 pKa = 10.16LGLSIIEE63 pKa = 4.36EE64 pKa = 4.51VNSHH68 pKa = 5.7IVVV71 pKa = 3.59
Molecular weight: 7.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P07946|SPIKE_CVPPU Spike glycoprotein OS=Porcine transmissible gastroenteritis coronavirus (strain Purdue) OX=11151 GN=S PE=1 SV=2
MM1 pKa = 7.48LVFLHH6 pKa = 6.78AVFITVLILLLIGRR20 pKa = 11.84LQLLEE25 pKa = 4.34RR26 pKa = 11.84LLLNHH31 pKa = 6.54SFNLKK36 pKa = 8.48TVNDD40 pKa = 3.73FNILYY45 pKa = 10.18RR46 pKa = 11.84SLAEE50 pKa = 3.86TRR52 pKa = 11.84LLKK55 pKa = 10.7VVLRR59 pKa = 11.84VIFLVLLGFCCYY71 pKa = 10.28RR72 pKa = 11.84LLVTLMM78 pKa = 4.2
MM1 pKa = 7.48LVFLHH6 pKa = 6.78AVFITVLILLLIGRR20 pKa = 11.84LQLLEE25 pKa = 4.34RR26 pKa = 11.84LLLNHH31 pKa = 6.54SFNLKK36 pKa = 8.48TVNDD40 pKa = 3.73FNILYY45 pKa = 10.18RR46 pKa = 11.84SLAEE50 pKa = 3.86TRR52 pKa = 11.84LLKK55 pKa = 10.7VVLRR59 pKa = 11.84VIFLVLLGFCCYY71 pKa = 10.28RR72 pKa = 11.84LLVTLMM78 pKa = 4.2
Molecular weight: 9.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13267 |
71 |
6684 |
1474.1 |
164.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.527 ± 0.186 | 3.136 ± 0.344 |
5.306 ± 0.211 | 4.914 ± 0.391 |
5.683 ± 0.245 | 6.422 ± 0.119 |
1.651 ± 0.114 | 5.992 ± 0.239 |
6.301 ± 0.78 | 8.238 ± 0.512 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.299 ± 0.28 | 6.075 ± 0.333 |
3.286 ± 0.11 | 2.925 ± 0.336 |
3.196 ± 0.585 | 6.904 ± 0.426 |
6.068 ± 0.542 | 9.218 ± 0.349 |
1.176 ± 0.205 | 4.681 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
