
Panthera leo polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
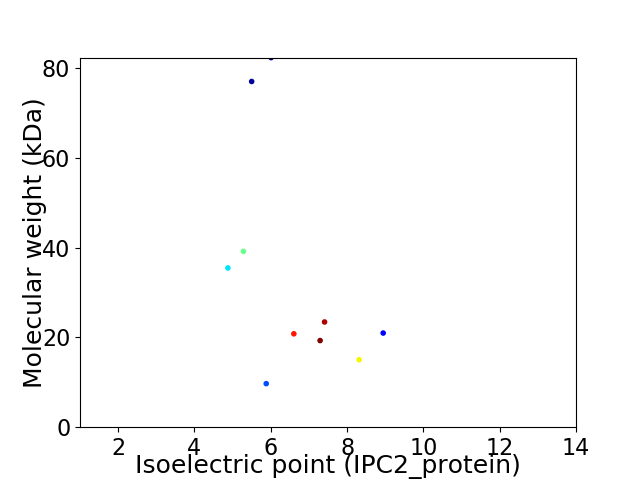
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S1CJP5|A0A2S1CJP5_9POLY 83T protein OS=Panthera leo polyomavirus 1 OX=2170405 GN=83T PE=4 SV=1
MM1 pKa = 7.46FFILGMGALLAVLAEE16 pKa = 4.28VFDD19 pKa = 4.55LATAAGVGIDD29 pKa = 4.51AILSGEE35 pKa = 4.16ALATAEE41 pKa = 4.06ALEE44 pKa = 4.3AFVTNLVTLEE54 pKa = 4.21GFTEE58 pKa = 4.44AEE60 pKa = 3.93ALAAAGITQEE70 pKa = 4.63ALTAFNQFGAVFPQLYY86 pKa = 9.98AALAVDD92 pKa = 4.34SVAIGSALAASLAPYY107 pKa = 9.29NYY109 pKa = 10.48SYY111 pKa = 7.65TTPIANLNMALIPYY125 pKa = 9.68IGEE128 pKa = 3.87LDD130 pKa = 3.41IYY132 pKa = 10.77FPGAMHH138 pKa = 7.42IARR141 pKa = 11.84FIHH144 pKa = 6.85YY145 pKa = 9.6IDD147 pKa = 3.75PTNWAVDD154 pKa = 4.29LYY156 pKa = 10.52QALGRR161 pKa = 11.84YY162 pKa = 7.21VWEE165 pKa = 4.45GLQRR169 pKa = 11.84AGQRR173 pKa = 11.84FLDD176 pKa = 3.78NQRR179 pKa = 11.84LQLEE183 pKa = 4.28HH184 pKa = 6.68ATRR187 pKa = 11.84EE188 pKa = 4.0LAQRR192 pKa = 11.84GAQTAAEE199 pKa = 4.04ALAQYY204 pKa = 10.03FEE206 pKa = 4.14NARR209 pKa = 11.84WAISNFNPQNLYY221 pKa = 10.59RR222 pKa = 11.84GLEE225 pKa = 3.74NYY227 pKa = 9.02YY228 pKa = 10.53RR229 pKa = 11.84QLPPLNPIQRR239 pKa = 11.84RR240 pKa = 11.84QLDD243 pKa = 3.49RR244 pKa = 11.84RR245 pKa = 11.84AGLPVPDD252 pKa = 3.66RR253 pKa = 11.84VLAIEE258 pKa = 5.16DD259 pKa = 3.73NVKK262 pKa = 10.54SADD265 pKa = 3.59YY266 pKa = 10.47VEE268 pKa = 6.05RR269 pKa = 11.84YY270 pKa = 9.36APPGGAFQRR279 pKa = 11.84QTPDD283 pKa = 2.11WMLPLILGLYY293 pKa = 10.41GDD295 pKa = 5.31INPSWGDD302 pKa = 3.24TLTQLEE308 pKa = 4.98KK309 pKa = 11.08EE310 pKa = 4.69DD311 pKa = 4.31GPPSKK316 pKa = 10.22KK317 pKa = 9.85RR318 pKa = 11.84KK319 pKa = 8.05KK320 pKa = 9.17TII322 pKa = 3.26
MM1 pKa = 7.46FFILGMGALLAVLAEE16 pKa = 4.28VFDD19 pKa = 4.55LATAAGVGIDD29 pKa = 4.51AILSGEE35 pKa = 4.16ALATAEE41 pKa = 4.06ALEE44 pKa = 4.3AFVTNLVTLEE54 pKa = 4.21GFTEE58 pKa = 4.44AEE60 pKa = 3.93ALAAAGITQEE70 pKa = 4.63ALTAFNQFGAVFPQLYY86 pKa = 9.98AALAVDD92 pKa = 4.34SVAIGSALAASLAPYY107 pKa = 9.29NYY109 pKa = 10.48SYY111 pKa = 7.65TTPIANLNMALIPYY125 pKa = 9.68IGEE128 pKa = 3.87LDD130 pKa = 3.41IYY132 pKa = 10.77FPGAMHH138 pKa = 7.42IARR141 pKa = 11.84FIHH144 pKa = 6.85YY145 pKa = 9.6IDD147 pKa = 3.75PTNWAVDD154 pKa = 4.29LYY156 pKa = 10.52QALGRR161 pKa = 11.84YY162 pKa = 7.21VWEE165 pKa = 4.45GLQRR169 pKa = 11.84AGQRR173 pKa = 11.84FLDD176 pKa = 3.78NQRR179 pKa = 11.84LQLEE183 pKa = 4.28HH184 pKa = 6.68ATRR187 pKa = 11.84EE188 pKa = 4.0LAQRR192 pKa = 11.84GAQTAAEE199 pKa = 4.04ALAQYY204 pKa = 10.03FEE206 pKa = 4.14NARR209 pKa = 11.84WAISNFNPQNLYY221 pKa = 10.59RR222 pKa = 11.84GLEE225 pKa = 3.74NYY227 pKa = 9.02YY228 pKa = 10.53RR229 pKa = 11.84QLPPLNPIQRR239 pKa = 11.84RR240 pKa = 11.84QLDD243 pKa = 3.49RR244 pKa = 11.84RR245 pKa = 11.84AGLPVPDD252 pKa = 3.66RR253 pKa = 11.84VLAIEE258 pKa = 5.16DD259 pKa = 3.73NVKK262 pKa = 10.54SADD265 pKa = 3.59YY266 pKa = 10.47VEE268 pKa = 6.05RR269 pKa = 11.84YY270 pKa = 9.36APPGGAFQRR279 pKa = 11.84QTPDD283 pKa = 2.11WMLPLILGLYY293 pKa = 10.41GDD295 pKa = 5.31INPSWGDD302 pKa = 3.24TLTQLEE308 pKa = 4.98KK309 pKa = 11.08EE310 pKa = 4.69DD311 pKa = 4.31GPPSKK316 pKa = 10.22KK317 pKa = 9.85RR318 pKa = 11.84KK319 pKa = 8.05KK320 pKa = 9.17TII322 pKa = 3.26
Molecular weight: 35.46 kDa
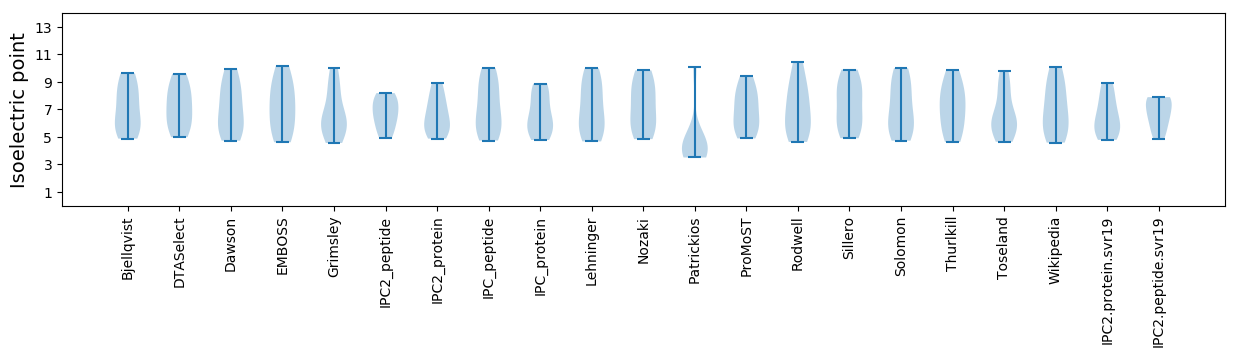
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S1CJQ6|A0A2S1CJQ6_9POLY 181T protein OS=Panthera leo polyomavirus 1 OX=2170405 GN=181T PE=4 SV=1
MM1 pKa = 7.6DD2 pKa = 6.14HH3 pKa = 7.11ILTRR7 pKa = 11.84EE8 pKa = 3.63EE9 pKa = 4.28SKK11 pKa = 11.2RR12 pKa = 11.84LMEE15 pKa = 4.97LLNLPMTEE23 pKa = 4.1YY24 pKa = 11.46GNFPLMRR31 pKa = 11.84RR32 pKa = 11.84AFLKK36 pKa = 10.24ACKK39 pKa = 9.45ILHH42 pKa = 6.55PDD44 pKa = 2.86KK45 pKa = 11.28GGNAEE50 pKa = 4.03QAQEE54 pKa = 4.5LISLYY59 pKa = 10.54RR60 pKa = 11.84KK61 pKa = 10.2LEE63 pKa = 3.91EE64 pKa = 4.73SLPSLNPQEE73 pKa = 5.69SFTTDD78 pKa = 2.75QKK80 pKa = 11.17KK81 pKa = 9.21QEE83 pKa = 4.1QEE85 pKa = 4.18TQGLGCMLLLRR96 pKa = 11.84MLYY99 pKa = 9.82SLVWAGIYY107 pKa = 9.23LLCLLLLDD115 pKa = 4.93GNNSQFTLWLLEE127 pKa = 3.83YY128 pKa = 10.12LVWKK132 pKa = 10.24ILMRR136 pKa = 11.84GNPNPAPASSKK147 pKa = 9.23EE148 pKa = 4.24EE149 pKa = 3.96KK150 pKa = 10.52GNPPMTLPPAPKK162 pKa = 8.91PRR164 pKa = 11.84HH165 pKa = 5.92RR166 pKa = 11.84KK167 pKa = 8.53SQLSKK172 pKa = 10.47KK173 pKa = 9.03KK174 pKa = 10.03KK175 pKa = 8.68KK176 pKa = 9.78SRR178 pKa = 11.84RR179 pKa = 11.84TWW181 pKa = 2.86
MM1 pKa = 7.6DD2 pKa = 6.14HH3 pKa = 7.11ILTRR7 pKa = 11.84EE8 pKa = 3.63EE9 pKa = 4.28SKK11 pKa = 11.2RR12 pKa = 11.84LMEE15 pKa = 4.97LLNLPMTEE23 pKa = 4.1YY24 pKa = 11.46GNFPLMRR31 pKa = 11.84RR32 pKa = 11.84AFLKK36 pKa = 10.24ACKK39 pKa = 9.45ILHH42 pKa = 6.55PDD44 pKa = 2.86KK45 pKa = 11.28GGNAEE50 pKa = 4.03QAQEE54 pKa = 4.5LISLYY59 pKa = 10.54RR60 pKa = 11.84KK61 pKa = 10.2LEE63 pKa = 3.91EE64 pKa = 4.73SLPSLNPQEE73 pKa = 5.69SFTTDD78 pKa = 2.75QKK80 pKa = 11.17KK81 pKa = 9.21QEE83 pKa = 4.1QEE85 pKa = 4.18TQGLGCMLLLRR96 pKa = 11.84MLYY99 pKa = 9.82SLVWAGIYY107 pKa = 9.23LLCLLLLDD115 pKa = 4.93GNNSQFTLWLLEE127 pKa = 3.83YY128 pKa = 10.12LVWKK132 pKa = 10.24ILMRR136 pKa = 11.84GNPNPAPASSKK147 pKa = 9.23EE148 pKa = 4.24EE149 pKa = 3.96KK150 pKa = 10.52GNPPMTLPPAPKK162 pKa = 8.91PRR164 pKa = 11.84HH165 pKa = 5.92RR166 pKa = 11.84KK167 pKa = 8.53SQLSKK172 pKa = 10.47KK173 pKa = 9.03KK174 pKa = 10.03KK175 pKa = 8.68KK176 pKa = 9.78SRR178 pKa = 11.84RR179 pKa = 11.84TWW181 pKa = 2.86
Molecular weight: 20.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3005 |
83 |
713 |
300.5 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.29 ± 1.034 | 2.762 ± 0.513 |
5.125 ± 0.409 | 7.488 ± 0.347 |
4.592 ± 0.447 | 5.358 ± 0.485 |
1.364 ± 0.148 | 5.125 ± 0.372 |
6.955 ± 0.749 | 11.78 ± 0.673 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.662 ± 0.342 | 4.992 ± 0.119 |
5.857 ± 0.663 | 4.759 ± 0.318 |
5.291 ± 0.347 | 5.691 ± 0.482 |
4.626 ± 0.269 | 4.393 ± 0.524 |
1.231 ± 0.167 | 3.661 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
