
Avian paramyxovirus goose/Shimane/67/2000
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Orthoavulavirus; Avian orthoavulavirus 13
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
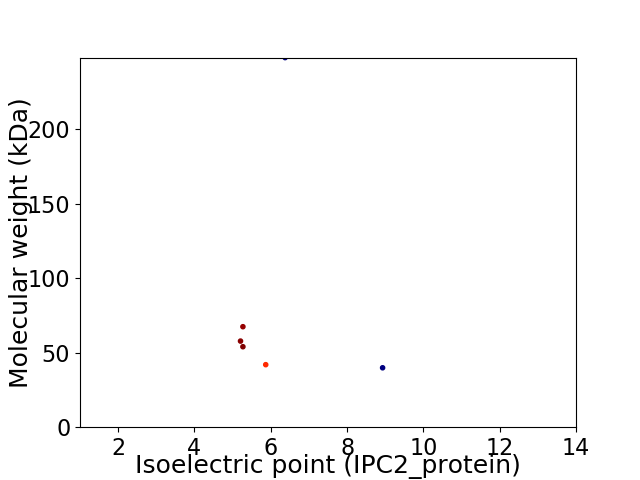
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A173M8Y4|A0A173M8Y4_9MONO Phosphoprotein OS=Avian paramyxovirus goose/Shimane/67/2000 OX=1401445 GN=P PE=4 SV=1
MM1 pKa = 7.54ARR3 pKa = 11.84FSQEE7 pKa = 3.2IFGLSTILLIAQTCQGSIDD26 pKa = 3.72GRR28 pKa = 11.84LTLAAGIVPVGDD40 pKa = 3.84RR41 pKa = 11.84PISIYY46 pKa = 10.3TSSQTGIIVVKK57 pKa = 9.71LIPNLPDD64 pKa = 3.3NKK66 pKa = 10.0KK67 pKa = 10.58DD68 pKa = 3.57CAKK71 pKa = 10.62QSLQSYY77 pKa = 10.07NEE79 pKa = 3.98TLSRR83 pKa = 11.84ILTPLATAMSAIRR96 pKa = 11.84GNSTTQVRR104 pKa = 11.84EE105 pKa = 3.7NRR107 pKa = 11.84LVGAIIGSVALGVATAAQITAATALIQANQNAANIARR144 pKa = 11.84LANSIAKK151 pKa = 8.97TNEE154 pKa = 3.37AVTDD158 pKa = 3.83LTEE161 pKa = 4.37GLGTLAIGVGKK172 pKa = 9.68LQDD175 pKa = 3.65YY176 pKa = 11.19VNEE179 pKa = 4.23QFNNTAVAIDD189 pKa = 4.04CLTLEE194 pKa = 4.32SRR196 pKa = 11.84LGIQLSLYY204 pKa = 8.73LTEE207 pKa = 4.34LMGVFGNQLTSPALTPITIQALYY230 pKa = 10.45NLAGGNLNALLSRR243 pKa = 11.84LGASEE248 pKa = 4.21TQLGSLINSGLIKK261 pKa = 10.44GMPIMYY267 pKa = 10.24DD268 pKa = 3.42DD269 pKa = 4.49ANKK272 pKa = 10.36LLAVQVEE279 pKa = 4.44LPSIGKK285 pKa = 9.81LNGARR290 pKa = 11.84STLLEE295 pKa = 4.14TLAVDD300 pKa = 4.06TTRR303 pKa = 11.84GPSSPIIPSAVIEE316 pKa = 4.24IGGAMEE322 pKa = 4.79EE323 pKa = 4.84LDD325 pKa = 4.8LSPCITTDD333 pKa = 4.04LDD335 pKa = 3.8MFCTKK340 pKa = 10.22IISYY344 pKa = 9.39PLSQSTLSCLNGNLSDD360 pKa = 4.19CVFSRR365 pKa = 11.84SEE367 pKa = 4.29GVLSTPYY374 pKa = 8.38MTIKK378 pKa = 10.66GKK380 pKa = 10.15IVANCKK386 pKa = 8.89QVICRR391 pKa = 11.84CMDD394 pKa = 3.97PPQILSQNYY403 pKa = 9.37GEE405 pKa = 4.89ALLLIDD411 pKa = 4.95EE412 pKa = 4.62STCRR416 pKa = 11.84SLEE419 pKa = 3.88LSGVILKK426 pKa = 10.35LAGTYY431 pKa = 8.9EE432 pKa = 4.1SEE434 pKa = 4.21YY435 pKa = 10.47TRR437 pKa = 11.84NLTVDD442 pKa = 4.12PSQVIITGPLDD453 pKa = 3.26ISAEE457 pKa = 3.98LSKK460 pKa = 11.13VNQSIDD466 pKa = 3.48SAKK469 pKa = 10.85EE470 pKa = 3.57NIAEE474 pKa = 4.3SNKK477 pKa = 9.86FLSQVNVKK485 pKa = 9.87LLSSSAMITYY495 pKa = 9.9IVATVVCLIIAITGCVIGIYY515 pKa = 9.81TLTKK519 pKa = 10.43LKK521 pKa = 10.53SQQEE525 pKa = 4.22TLLWLGNNAEE535 pKa = 4.31MHH537 pKa = 6.49GSRR540 pKa = 11.84SKK542 pKa = 10.13TSFF545 pKa = 2.94
MM1 pKa = 7.54ARR3 pKa = 11.84FSQEE7 pKa = 3.2IFGLSTILLIAQTCQGSIDD26 pKa = 3.72GRR28 pKa = 11.84LTLAAGIVPVGDD40 pKa = 3.84RR41 pKa = 11.84PISIYY46 pKa = 10.3TSSQTGIIVVKK57 pKa = 9.71LIPNLPDD64 pKa = 3.3NKK66 pKa = 10.0KK67 pKa = 10.58DD68 pKa = 3.57CAKK71 pKa = 10.62QSLQSYY77 pKa = 10.07NEE79 pKa = 3.98TLSRR83 pKa = 11.84ILTPLATAMSAIRR96 pKa = 11.84GNSTTQVRR104 pKa = 11.84EE105 pKa = 3.7NRR107 pKa = 11.84LVGAIIGSVALGVATAAQITAATALIQANQNAANIARR144 pKa = 11.84LANSIAKK151 pKa = 8.97TNEE154 pKa = 3.37AVTDD158 pKa = 3.83LTEE161 pKa = 4.37GLGTLAIGVGKK172 pKa = 9.68LQDD175 pKa = 3.65YY176 pKa = 11.19VNEE179 pKa = 4.23QFNNTAVAIDD189 pKa = 4.04CLTLEE194 pKa = 4.32SRR196 pKa = 11.84LGIQLSLYY204 pKa = 8.73LTEE207 pKa = 4.34LMGVFGNQLTSPALTPITIQALYY230 pKa = 10.45NLAGGNLNALLSRR243 pKa = 11.84LGASEE248 pKa = 4.21TQLGSLINSGLIKK261 pKa = 10.44GMPIMYY267 pKa = 10.24DD268 pKa = 3.42DD269 pKa = 4.49ANKK272 pKa = 10.36LLAVQVEE279 pKa = 4.44LPSIGKK285 pKa = 9.81LNGARR290 pKa = 11.84STLLEE295 pKa = 4.14TLAVDD300 pKa = 4.06TTRR303 pKa = 11.84GPSSPIIPSAVIEE316 pKa = 4.24IGGAMEE322 pKa = 4.79EE323 pKa = 4.84LDD325 pKa = 4.8LSPCITTDD333 pKa = 4.04LDD335 pKa = 3.8MFCTKK340 pKa = 10.22IISYY344 pKa = 9.39PLSQSTLSCLNGNLSDD360 pKa = 4.19CVFSRR365 pKa = 11.84SEE367 pKa = 4.29GVLSTPYY374 pKa = 8.38MTIKK378 pKa = 10.66GKK380 pKa = 10.15IVANCKK386 pKa = 8.89QVICRR391 pKa = 11.84CMDD394 pKa = 3.97PPQILSQNYY403 pKa = 9.37GEE405 pKa = 4.89ALLLIDD411 pKa = 4.95EE412 pKa = 4.62STCRR416 pKa = 11.84SLEE419 pKa = 3.88LSGVILKK426 pKa = 10.35LAGTYY431 pKa = 8.9EE432 pKa = 4.1SEE434 pKa = 4.21YY435 pKa = 10.47TRR437 pKa = 11.84NLTVDD442 pKa = 4.12PSQVIITGPLDD453 pKa = 3.26ISAEE457 pKa = 3.98LSKK460 pKa = 11.13VNQSIDD466 pKa = 3.48SAKK469 pKa = 10.85EE470 pKa = 3.57NIAEE474 pKa = 4.3SNKK477 pKa = 9.86FLSQVNVKK485 pKa = 9.87LLSSSAMITYY495 pKa = 9.9IVATVVCLIIAITGCVIGIYY515 pKa = 9.81TLTKK519 pKa = 10.43LKK521 pKa = 10.53SQQEE525 pKa = 4.22TLLWLGNNAEE535 pKa = 4.31MHH537 pKa = 6.49GSRR540 pKa = 11.84SKK542 pKa = 10.13TSFF545 pKa = 2.94
Molecular weight: 57.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A173M8X5|A0A173M8X5_9MONO Hemagglutinin-neuraminidase OS=Avian paramyxovirus goose/Shimane/67/2000 OX=1401445 GN=HN PE=3 SV=1
MM1 pKa = 8.06DD2 pKa = 4.14SSKK5 pKa = 10.76IIGLEE10 pKa = 3.76VDD12 pKa = 3.97PSSPSNTLLAFPVVLQEE29 pKa = 3.67IDD31 pKa = 3.31GGRR34 pKa = 11.84KK35 pKa = 9.37EE36 pKa = 3.83ITPQFRR42 pKa = 11.84TQKK45 pKa = 9.73IDD47 pKa = 2.81MWSEE51 pKa = 4.16SKK53 pKa = 10.64SDD55 pKa = 3.54SVSITTYY62 pKa = 10.55GFIYY66 pKa = 9.87GVKK69 pKa = 10.14GGNDD73 pKa = 2.97NSGPIMAEE81 pKa = 3.8QQKK84 pKa = 9.87EE85 pKa = 4.11PLSAAMLCFGSVGYY99 pKa = 10.64NSGLPEE105 pKa = 3.83IARR108 pKa = 11.84AALNTIITCKK118 pKa = 10.37KK119 pKa = 9.7SATDD123 pKa = 3.3SEE125 pKa = 5.04RR126 pKa = 11.84ILFTVHH132 pKa = 6.16QAPQMIQEE140 pKa = 4.51AKK142 pKa = 10.4VISTRR147 pKa = 11.84YY148 pKa = 10.02SSVAANKK155 pKa = 9.81CVRR158 pKa = 11.84APEE161 pKa = 4.19RR162 pKa = 11.84VPSGLSLEE170 pKa = 4.41YY171 pKa = 10.53KK172 pKa = 9.15VTFVSLTVVPKK183 pKa = 10.13SDD185 pKa = 3.36VYY187 pKa = 10.77KK188 pKa = 10.69VPRR191 pKa = 11.84PVLRR195 pKa = 11.84LHH197 pKa = 6.54SKK199 pKa = 10.12HH200 pKa = 6.16VLNIAINAVIQIDD213 pKa = 3.67IQATHH218 pKa = 7.39PLAKK222 pKa = 9.84TLIKK226 pKa = 10.91RR227 pKa = 11.84NDD229 pKa = 3.29QFFADD234 pKa = 4.46LFIHH238 pKa = 6.99IGMISCLDD246 pKa = 3.51SKK248 pKa = 11.23GNKK251 pKa = 9.14ISVDD255 pKa = 3.22KK256 pKa = 11.49LEE258 pKa = 4.15LKK260 pKa = 9.67IRR262 pKa = 11.84RR263 pKa = 11.84MSISVGLLDD272 pKa = 4.76IFGPSIALKK281 pKa = 10.78ARR283 pKa = 11.84GKK285 pKa = 8.16RR286 pKa = 11.84TKK288 pKa = 10.64VMSPFFSPRR297 pKa = 11.84GTACYY302 pKa = 9.21PISQTAPGIAKK313 pKa = 9.82ILWSQTGSLHH323 pKa = 5.01EE324 pKa = 4.57CKK326 pKa = 10.24IIIQGGTNRR335 pKa = 11.84AIATTDD341 pKa = 3.4DD342 pKa = 4.02FVVGSTKK349 pKa = 9.67IEE351 pKa = 3.77KK352 pKa = 10.06SGRR355 pKa = 11.84NGKK358 pKa = 9.62FNPFKK363 pKa = 10.55KK364 pKa = 9.16TGG366 pKa = 3.25
MM1 pKa = 8.06DD2 pKa = 4.14SSKK5 pKa = 10.76IIGLEE10 pKa = 3.76VDD12 pKa = 3.97PSSPSNTLLAFPVVLQEE29 pKa = 3.67IDD31 pKa = 3.31GGRR34 pKa = 11.84KK35 pKa = 9.37EE36 pKa = 3.83ITPQFRR42 pKa = 11.84TQKK45 pKa = 9.73IDD47 pKa = 2.81MWSEE51 pKa = 4.16SKK53 pKa = 10.64SDD55 pKa = 3.54SVSITTYY62 pKa = 10.55GFIYY66 pKa = 9.87GVKK69 pKa = 10.14GGNDD73 pKa = 2.97NSGPIMAEE81 pKa = 3.8QQKK84 pKa = 9.87EE85 pKa = 4.11PLSAAMLCFGSVGYY99 pKa = 10.64NSGLPEE105 pKa = 3.83IARR108 pKa = 11.84AALNTIITCKK118 pKa = 10.37KK119 pKa = 9.7SATDD123 pKa = 3.3SEE125 pKa = 5.04RR126 pKa = 11.84ILFTVHH132 pKa = 6.16QAPQMIQEE140 pKa = 4.51AKK142 pKa = 10.4VISTRR147 pKa = 11.84YY148 pKa = 10.02SSVAANKK155 pKa = 9.81CVRR158 pKa = 11.84APEE161 pKa = 4.19RR162 pKa = 11.84VPSGLSLEE170 pKa = 4.41YY171 pKa = 10.53KK172 pKa = 9.15VTFVSLTVVPKK183 pKa = 10.13SDD185 pKa = 3.36VYY187 pKa = 10.77KK188 pKa = 10.69VPRR191 pKa = 11.84PVLRR195 pKa = 11.84LHH197 pKa = 6.54SKK199 pKa = 10.12HH200 pKa = 6.16VLNIAINAVIQIDD213 pKa = 3.67IQATHH218 pKa = 7.39PLAKK222 pKa = 9.84TLIKK226 pKa = 10.91RR227 pKa = 11.84NDD229 pKa = 3.29QFFADD234 pKa = 4.46LFIHH238 pKa = 6.99IGMISCLDD246 pKa = 3.51SKK248 pKa = 11.23GNKK251 pKa = 9.14ISVDD255 pKa = 3.22KK256 pKa = 11.49LEE258 pKa = 4.15LKK260 pKa = 9.67IRR262 pKa = 11.84RR263 pKa = 11.84MSISVGLLDD272 pKa = 4.76IFGPSIALKK281 pKa = 10.78ARR283 pKa = 11.84GKK285 pKa = 8.16RR286 pKa = 11.84TKK288 pKa = 10.64VMSPFFSPRR297 pKa = 11.84GTACYY302 pKa = 9.21PISQTAPGIAKK313 pKa = 9.82ILWSQTGSLHH323 pKa = 5.01EE324 pKa = 4.57CKK326 pKa = 10.24IIIQGGTNRR335 pKa = 11.84AIATTDD341 pKa = 3.4DD342 pKa = 4.02FVVGSTKK349 pKa = 9.67IEE351 pKa = 3.77KK352 pKa = 10.06SGRR355 pKa = 11.84NGKK358 pKa = 9.62FNPFKK363 pKa = 10.55KK364 pKa = 9.16TGG366 pKa = 3.25
Molecular weight: 39.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4610 |
366 |
2199 |
768.3 |
84.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.028 ± 0.719 | 1.931 ± 0.228 |
5.293 ± 0.313 | 5.054 ± 0.214 |
3.427 ± 0.456 | 6.334 ± 0.527 |
1.8 ± 0.389 | 8.156 ± 0.543 |
5.184 ± 0.486 | 10.087 ± 0.938 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.777 ± 0.282 | 4.989 ± 0.239 |
4.382 ± 0.433 | 4.121 ± 0.252 |
4.772 ± 0.393 | 9.111 ± 0.825 |
6.226 ± 0.541 | 5.466 ± 0.276 |
0.868 ± 0.152 | 2.993 ± 0.32 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
