
Cactus virus X
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Potexvirus
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
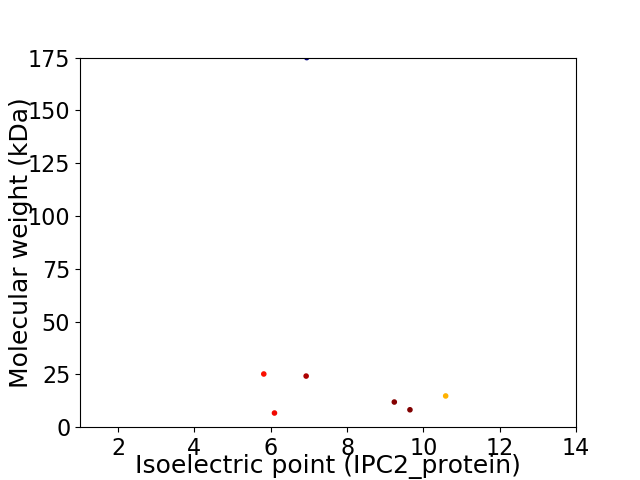
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91ND7|Q91ND7_9VIRU 8 kDa protein OS=Cactus virus X OX=112227 PE=4 SV=1
MM1 pKa = 7.01EE2 pKa = 5.12TLTALLIANGYY13 pKa = 6.89TRR15 pKa = 11.84TDD17 pKa = 3.59LPISKK22 pKa = 9.24PLVIHH27 pKa = 6.62AVAGAGKK34 pKa = 6.7TTLIRR39 pKa = 11.84QFLHH43 pKa = 4.98QHH45 pKa = 5.7SATNAQTLGTPDD57 pKa = 3.77KK58 pKa = 10.51PNLSRR63 pKa = 11.84KK64 pKa = 8.56MIRR67 pKa = 11.84QFSMPKK73 pKa = 9.88ANHH76 pKa = 5.96FNILDD81 pKa = 4.02EE82 pKa = 4.59YY83 pKa = 10.65CAQPLKK89 pKa = 10.81GSWDD93 pKa = 3.48AVFADD98 pKa = 4.67PLQHH102 pKa = 7.14PDD104 pKa = 3.32YY105 pKa = 10.84ALEE108 pKa = 3.72PHH110 pKa = 6.98FIKK113 pKa = 9.95EE114 pKa = 4.2TSHH117 pKa = 6.77RR118 pKa = 11.84LGPSTCEE125 pKa = 4.43LISSLGILIYY135 pKa = 10.08PDD137 pKa = 4.64SEE139 pKa = 4.43DD140 pKa = 3.44QVVTRR145 pKa = 11.84KK146 pKa = 10.15GVFEE150 pKa = 4.32SEE152 pKa = 4.09LFGVVIALDD161 pKa = 3.53EE162 pKa = 5.56AIFQLASRR170 pKa = 11.84HH171 pKa = 4.87GLKK174 pKa = 10.13PLCPKK179 pKa = 9.66ATIGLQFPVVTVLSSLALEE198 pKa = 4.11QVEE201 pKa = 5.23DD202 pKa = 3.69STALYY207 pKa = 10.04IALTRR212 pKa = 11.84HH213 pKa = 4.96TKK215 pKa = 9.25EE216 pKa = 3.6LHH218 pKa = 5.31VRR220 pKa = 11.84CPTEE224 pKa = 4.8ADD226 pKa = 3.08ATTT229 pKa = 3.47
MM1 pKa = 7.01EE2 pKa = 5.12TLTALLIANGYY13 pKa = 6.89TRR15 pKa = 11.84TDD17 pKa = 3.59LPISKK22 pKa = 9.24PLVIHH27 pKa = 6.62AVAGAGKK34 pKa = 6.7TTLIRR39 pKa = 11.84QFLHH43 pKa = 4.98QHH45 pKa = 5.7SATNAQTLGTPDD57 pKa = 3.77KK58 pKa = 10.51PNLSRR63 pKa = 11.84KK64 pKa = 8.56MIRR67 pKa = 11.84QFSMPKK73 pKa = 9.88ANHH76 pKa = 5.96FNILDD81 pKa = 4.02EE82 pKa = 4.59YY83 pKa = 10.65CAQPLKK89 pKa = 10.81GSWDD93 pKa = 3.48AVFADD98 pKa = 4.67PLQHH102 pKa = 7.14PDD104 pKa = 3.32YY105 pKa = 10.84ALEE108 pKa = 3.72PHH110 pKa = 6.98FIKK113 pKa = 9.95EE114 pKa = 4.2TSHH117 pKa = 6.77RR118 pKa = 11.84LGPSTCEE125 pKa = 4.43LISSLGILIYY135 pKa = 10.08PDD137 pKa = 4.64SEE139 pKa = 4.43DD140 pKa = 3.44QVVTRR145 pKa = 11.84KK146 pKa = 10.15GVFEE150 pKa = 4.32SEE152 pKa = 4.09LFGVVIALDD161 pKa = 3.53EE162 pKa = 5.56AIFQLASRR170 pKa = 11.84HH171 pKa = 4.87GLKK174 pKa = 10.13PLCPKK179 pKa = 9.66ATIGLQFPVVTVLSSLALEE198 pKa = 4.11QVEE201 pKa = 5.23DD202 pKa = 3.69STALYY207 pKa = 10.04IALTRR212 pKa = 11.84HH213 pKa = 4.96TKK215 pKa = 9.25EE216 pKa = 3.6LHH218 pKa = 5.31VRR220 pKa = 11.84CPTEE224 pKa = 4.8ADD226 pKa = 3.08ATTT229 pKa = 3.47
Molecular weight: 25.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91ND8|Q91ND8_9VIRU 15 kDa protein OS=Cactus virus X OX=112227 PE=4 SV=1
MM1 pKa = 7.59PHH3 pKa = 6.32TATGRR8 pKa = 11.84TWRR11 pKa = 11.84TCSEE15 pKa = 3.89STQRR19 pKa = 11.84LRR21 pKa = 11.84DD22 pKa = 3.66APTSPWTPPFFPAGICSSHH41 pKa = 6.04PCSRR45 pKa = 11.84NKK47 pKa = 9.02HH48 pKa = 4.87TGSWDD53 pKa = 3.21TRR55 pKa = 11.84FQPTPGVKK63 pKa = 9.43EE64 pKa = 4.05SRR66 pKa = 11.84RR67 pKa = 11.84MRR69 pKa = 11.84SKK71 pKa = 10.72YY72 pKa = 10.54
MM1 pKa = 7.59PHH3 pKa = 6.32TATGRR8 pKa = 11.84TWRR11 pKa = 11.84TCSEE15 pKa = 3.89STQRR19 pKa = 11.84LRR21 pKa = 11.84DD22 pKa = 3.66APTSPWTPPFFPAGICSSHH41 pKa = 6.04PCSRR45 pKa = 11.84NKK47 pKa = 9.02HH48 pKa = 4.87TGSWDD53 pKa = 3.21TRR55 pKa = 11.84FQPTPGVKK63 pKa = 9.43EE64 pKa = 4.05SRR66 pKa = 11.84RR67 pKa = 11.84MRR69 pKa = 11.84SKK71 pKa = 10.72YY72 pKa = 10.54
Molecular weight: 8.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2376 |
64 |
1543 |
339.4 |
38.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.987 ± 0.53 | 1.557 ± 0.254 |
4.167 ± 0.698 | 5.303 ± 0.879 |
4.63 ± 0.538 | 5.429 ± 0.415 |
3.62 ± 0.389 | 5.219 ± 0.709 |
6.061 ± 1.071 | 10.101 ± 1.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.395 | 4.125 ± 0.474 |
7.323 ± 0.835 | 4.335 ± 0.67 |
4.714 ± 0.951 | 7.786 ± 1.482 |
7.997 ± 0.736 | 4.461 ± 0.558 |
1.473 ± 0.237 | 2.483 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
