
Mariniphaga sediminis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Mariniphaga
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
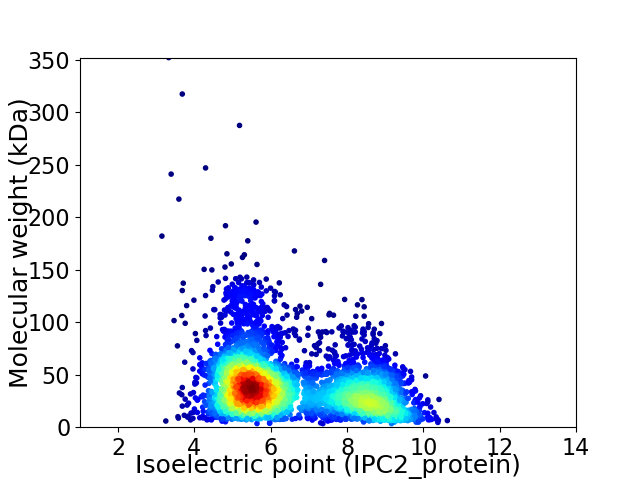
Virtual 2D-PAGE plot for 4517 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A399D6A9|A0A399D6A9_9BACT Dipeptidase OS=Mariniphaga sediminis OX=1628158 GN=D1164_00970 PE=3 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.36IYY5 pKa = 10.62LLLLIMIYY13 pKa = 10.48SSALMAQDD21 pKa = 3.9CPGDD25 pKa = 3.26IVVNNDD31 pKa = 3.41PGICGAVVDD40 pKa = 4.67YY41 pKa = 10.36AAPSPAGATVTQIDD55 pKa = 4.19EE56 pKa = 4.47SGLTSGDD63 pKa = 3.43QFPVGEE69 pKa = 4.2TLQEE73 pKa = 3.98YY74 pKa = 10.13EE75 pKa = 4.21LDD77 pKa = 3.7YY78 pKa = 11.62GIGGKK83 pKa = 7.58DD84 pKa = 2.9TCSFTVTVNDD94 pKa = 3.74TEE96 pKa = 5.02APLLSCQNISVNNDD110 pKa = 3.14PGEE113 pKa = 4.4AGAEE117 pKa = 4.03VTIPVPYY124 pKa = 9.94HH125 pKa = 5.92NNPAVTEE132 pKa = 4.29TVDD135 pKa = 4.21VLLLYY140 pKa = 10.67ADD142 pKa = 4.13YY143 pKa = 11.08DD144 pKa = 4.07SYY146 pKa = 11.97AQDD149 pKa = 3.14VKK151 pKa = 11.47AKK153 pKa = 10.72LLGTGNFSTVHH164 pKa = 5.83TFNISSRR171 pKa = 11.84VPEE174 pKa = 4.2LEE176 pKa = 3.87GLLEE180 pKa = 4.32FDD182 pKa = 6.01VIMLWRR188 pKa = 11.84NGSFPDD194 pKa = 4.11PEE196 pKa = 4.87ALGNVLAGFVDD207 pKa = 3.95AGRR210 pKa = 11.84GLVMAGNALSDD221 pKa = 3.19ISYY224 pKa = 10.14LAGAFEE230 pKa = 4.08TDD232 pKa = 3.34EE233 pKa = 4.54YY234 pKa = 11.51GVIVPGYY241 pKa = 8.49IQYY244 pKa = 10.81GGQAYY249 pKa = 10.47LGDD252 pKa = 3.91VHH254 pKa = 6.47LTSHH258 pKa = 7.34PLMNSITSFDD268 pKa = 3.88GGLLSYY274 pKa = 10.69RR275 pKa = 11.84SISNAITPGSYY286 pKa = 9.26RR287 pKa = 11.84VADD290 pKa = 3.63WTDD293 pKa = 3.47GNPLIAAKK301 pKa = 10.41EE302 pKa = 3.9NVGASNARR310 pKa = 11.84RR311 pKa = 11.84VDD313 pKa = 3.98LNFFPPTVTDD323 pKa = 3.56HH324 pKa = 6.9GRR326 pKa = 11.84VPWDD330 pKa = 3.32VNTDD334 pKa = 2.88GDD336 pKa = 5.1LIMANAILWVAGAPIAYY353 pKa = 8.45DD354 pKa = 2.98NCGPVEE360 pKa = 4.05FSNNFNASTSPSGYY374 pKa = 9.97YY375 pKa = 9.93PIGSTSVTLNATDD388 pKa = 3.62EE389 pKa = 4.55AGNTSTCNFTVTVEE403 pKa = 5.21DD404 pKa = 3.7IDD406 pKa = 4.37PPVAICKK413 pKa = 10.07NISVVLDD420 pKa = 3.52EE421 pKa = 4.98NGQVTIEE428 pKa = 4.25GSDD431 pKa = 3.53VDD433 pKa = 5.24GGSTDD438 pKa = 2.98NHH440 pKa = 6.77AIATYY445 pKa = 10.44EE446 pKa = 4.11LDD448 pKa = 3.52TNTFDD453 pKa = 6.54DD454 pKa = 4.84SMLGEE459 pKa = 4.22NTVVLTVIDD468 pKa = 3.81EE469 pKa = 4.75AGNSSTCTATVQVFDD484 pKa = 4.88NNPPDD489 pKa = 4.07IVVQPIEE496 pKa = 4.07VALDD500 pKa = 3.24NTGNYY505 pKa = 9.31EE506 pKa = 4.3LTLEE510 pKa = 4.59DD511 pKa = 4.28LKK513 pKa = 11.6EE514 pKa = 4.08MAEE517 pKa = 4.2GTTDD521 pKa = 3.25NGTSFDD527 pKa = 3.84NLKK530 pKa = 10.64FSAFPTTFACEE541 pKa = 4.11NVGEE545 pKa = 4.41EE546 pKa = 3.94INVRR550 pKa = 11.84LTVEE554 pKa = 4.33DD555 pKa = 3.95EE556 pKa = 4.49SGNSDD561 pKa = 3.73KK562 pKa = 11.02KK563 pKa = 7.79WTSVTVVDD571 pKa = 5.53IIPPTVVCQNIEE583 pKa = 3.98VYY585 pKa = 10.82LDD587 pKa = 3.6EE588 pKa = 5.64NGEE591 pKa = 3.91ASITPEE597 pKa = 4.27EE598 pKa = 4.34INVSGEE604 pKa = 3.77GGSYY608 pKa = 9.76DD609 pKa = 3.24ACGIASLEE617 pKa = 4.03LDD619 pKa = 3.52KK620 pKa = 11.06MAFGCEE626 pKa = 3.82DD627 pKa = 3.11TGEE630 pKa = 4.28NNVVLSVTDD639 pKa = 3.34HH640 pKa = 6.46GGNTGSCTAIVTVKK654 pKa = 10.07DD655 pKa = 3.23TFPPVFEE662 pKa = 4.26PVADD666 pKa = 3.01IVVEE670 pKa = 4.1VEE672 pKa = 4.11TNITEE677 pKa = 4.27TTIDD681 pKa = 3.58YY682 pKa = 10.16PNIVVTDD689 pKa = 3.71NCSVTLEE696 pKa = 4.21LTEE699 pKa = 4.26GLGPDD704 pKa = 3.48GVFPVGTTTEE714 pKa = 3.67TWAASDD720 pKa = 3.63PAGNSMTLTFTVTVTPVNSPPVLVNPIADD749 pKa = 3.51QAVNASYY756 pKa = 10.86VLKK759 pKa = 10.95VPVRR763 pKa = 11.84PEE765 pKa = 3.67LGEE768 pKa = 4.07VFDD771 pKa = 6.16DD772 pKa = 4.06VDD774 pKa = 4.05GDD776 pKa = 4.02EE777 pKa = 4.44LTISAMLEE785 pKa = 4.01NGDD788 pKa = 5.03PLPAWAEE795 pKa = 4.05MVDD798 pKa = 3.83DD799 pKa = 4.55SLVFTPLIADD809 pKa = 4.3TGCVSIIVKK818 pKa = 8.99ATDD821 pKa = 3.52PDD823 pKa = 3.71GAAAADD829 pKa = 3.98TFQLCVDD836 pKa = 4.97GYY838 pKa = 9.14PVSSPAVDD846 pKa = 3.33AAALNVTLYY855 pKa = 10.09PNPTRR860 pKa = 11.84DD861 pKa = 2.99RR862 pKa = 11.84VNIEE866 pKa = 3.9VQNSISGPAEE876 pKa = 3.31ISVFTMDD883 pKa = 4.8GKK885 pKa = 11.01RR886 pKa = 11.84ILQRR890 pKa = 11.84NYY892 pKa = 8.57TDD894 pKa = 2.87NRR896 pKa = 11.84RR897 pKa = 11.84ISFSMKK903 pKa = 8.23EE904 pKa = 4.39HH905 pKa = 5.58VSGMYY910 pKa = 9.26FVKK913 pKa = 10.79LNMQGKK919 pKa = 9.23EE920 pKa = 3.89IVKK923 pKa = 10.6KK924 pKa = 10.71LVLNNKK930 pKa = 8.99
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.36IYY5 pKa = 10.62LLLLIMIYY13 pKa = 10.48SSALMAQDD21 pKa = 3.9CPGDD25 pKa = 3.26IVVNNDD31 pKa = 3.41PGICGAVVDD40 pKa = 4.67YY41 pKa = 10.36AAPSPAGATVTQIDD55 pKa = 4.19EE56 pKa = 4.47SGLTSGDD63 pKa = 3.43QFPVGEE69 pKa = 4.2TLQEE73 pKa = 3.98YY74 pKa = 10.13EE75 pKa = 4.21LDD77 pKa = 3.7YY78 pKa = 11.62GIGGKK83 pKa = 7.58DD84 pKa = 2.9TCSFTVTVNDD94 pKa = 3.74TEE96 pKa = 5.02APLLSCQNISVNNDD110 pKa = 3.14PGEE113 pKa = 4.4AGAEE117 pKa = 4.03VTIPVPYY124 pKa = 9.94HH125 pKa = 5.92NNPAVTEE132 pKa = 4.29TVDD135 pKa = 4.21VLLLYY140 pKa = 10.67ADD142 pKa = 4.13YY143 pKa = 11.08DD144 pKa = 4.07SYY146 pKa = 11.97AQDD149 pKa = 3.14VKK151 pKa = 11.47AKK153 pKa = 10.72LLGTGNFSTVHH164 pKa = 5.83TFNISSRR171 pKa = 11.84VPEE174 pKa = 4.2LEE176 pKa = 3.87GLLEE180 pKa = 4.32FDD182 pKa = 6.01VIMLWRR188 pKa = 11.84NGSFPDD194 pKa = 4.11PEE196 pKa = 4.87ALGNVLAGFVDD207 pKa = 3.95AGRR210 pKa = 11.84GLVMAGNALSDD221 pKa = 3.19ISYY224 pKa = 10.14LAGAFEE230 pKa = 4.08TDD232 pKa = 3.34EE233 pKa = 4.54YY234 pKa = 11.51GVIVPGYY241 pKa = 8.49IQYY244 pKa = 10.81GGQAYY249 pKa = 10.47LGDD252 pKa = 3.91VHH254 pKa = 6.47LTSHH258 pKa = 7.34PLMNSITSFDD268 pKa = 3.88GGLLSYY274 pKa = 10.69RR275 pKa = 11.84SISNAITPGSYY286 pKa = 9.26RR287 pKa = 11.84VADD290 pKa = 3.63WTDD293 pKa = 3.47GNPLIAAKK301 pKa = 10.41EE302 pKa = 3.9NVGASNARR310 pKa = 11.84RR311 pKa = 11.84VDD313 pKa = 3.98LNFFPPTVTDD323 pKa = 3.56HH324 pKa = 6.9GRR326 pKa = 11.84VPWDD330 pKa = 3.32VNTDD334 pKa = 2.88GDD336 pKa = 5.1LIMANAILWVAGAPIAYY353 pKa = 8.45DD354 pKa = 2.98NCGPVEE360 pKa = 4.05FSNNFNASTSPSGYY374 pKa = 9.97YY375 pKa = 9.93PIGSTSVTLNATDD388 pKa = 3.62EE389 pKa = 4.55AGNTSTCNFTVTVEE403 pKa = 5.21DD404 pKa = 3.7IDD406 pKa = 4.37PPVAICKK413 pKa = 10.07NISVVLDD420 pKa = 3.52EE421 pKa = 4.98NGQVTIEE428 pKa = 4.25GSDD431 pKa = 3.53VDD433 pKa = 5.24GGSTDD438 pKa = 2.98NHH440 pKa = 6.77AIATYY445 pKa = 10.44EE446 pKa = 4.11LDD448 pKa = 3.52TNTFDD453 pKa = 6.54DD454 pKa = 4.84SMLGEE459 pKa = 4.22NTVVLTVIDD468 pKa = 3.81EE469 pKa = 4.75AGNSSTCTATVQVFDD484 pKa = 4.88NNPPDD489 pKa = 4.07IVVQPIEE496 pKa = 4.07VALDD500 pKa = 3.24NTGNYY505 pKa = 9.31EE506 pKa = 4.3LTLEE510 pKa = 4.59DD511 pKa = 4.28LKK513 pKa = 11.6EE514 pKa = 4.08MAEE517 pKa = 4.2GTTDD521 pKa = 3.25NGTSFDD527 pKa = 3.84NLKK530 pKa = 10.64FSAFPTTFACEE541 pKa = 4.11NVGEE545 pKa = 4.41EE546 pKa = 3.94INVRR550 pKa = 11.84LTVEE554 pKa = 4.33DD555 pKa = 3.95EE556 pKa = 4.49SGNSDD561 pKa = 3.73KK562 pKa = 11.02KK563 pKa = 7.79WTSVTVVDD571 pKa = 5.53IIPPTVVCQNIEE583 pKa = 3.98VYY585 pKa = 10.82LDD587 pKa = 3.6EE588 pKa = 5.64NGEE591 pKa = 3.91ASITPEE597 pKa = 4.27EE598 pKa = 4.34INVSGEE604 pKa = 3.77GGSYY608 pKa = 9.76DD609 pKa = 3.24ACGIASLEE617 pKa = 4.03LDD619 pKa = 3.52KK620 pKa = 11.06MAFGCEE626 pKa = 3.82DD627 pKa = 3.11TGEE630 pKa = 4.28NNVVLSVTDD639 pKa = 3.34HH640 pKa = 6.46GGNTGSCTAIVTVKK654 pKa = 10.07DD655 pKa = 3.23TFPPVFEE662 pKa = 4.26PVADD666 pKa = 3.01IVVEE670 pKa = 4.1VEE672 pKa = 4.11TNITEE677 pKa = 4.27TTIDD681 pKa = 3.58YY682 pKa = 10.16PNIVVTDD689 pKa = 3.71NCSVTLEE696 pKa = 4.21LTEE699 pKa = 4.26GLGPDD704 pKa = 3.48GVFPVGTTTEE714 pKa = 3.67TWAASDD720 pKa = 3.63PAGNSMTLTFTVTVTPVNSPPVLVNPIADD749 pKa = 3.51QAVNASYY756 pKa = 10.86VLKK759 pKa = 10.95VPVRR763 pKa = 11.84PEE765 pKa = 3.67LGEE768 pKa = 4.07VFDD771 pKa = 6.16DD772 pKa = 4.06VDD774 pKa = 4.05GDD776 pKa = 4.02EE777 pKa = 4.44LTISAMLEE785 pKa = 4.01NGDD788 pKa = 5.03PLPAWAEE795 pKa = 4.05MVDD798 pKa = 3.83DD799 pKa = 4.55SLVFTPLIADD809 pKa = 4.3TGCVSIIVKK818 pKa = 8.99ATDD821 pKa = 3.52PDD823 pKa = 3.71GAAAADD829 pKa = 3.98TFQLCVDD836 pKa = 4.97GYY838 pKa = 9.14PVSSPAVDD846 pKa = 3.33AAALNVTLYY855 pKa = 10.09PNPTRR860 pKa = 11.84DD861 pKa = 2.99RR862 pKa = 11.84VNIEE866 pKa = 3.9VQNSISGPAEE876 pKa = 3.31ISVFTMDD883 pKa = 4.8GKK885 pKa = 11.01RR886 pKa = 11.84ILQRR890 pKa = 11.84NYY892 pKa = 8.57TDD894 pKa = 2.87NRR896 pKa = 11.84RR897 pKa = 11.84ISFSMKK903 pKa = 8.23EE904 pKa = 4.39HH905 pKa = 5.58VSGMYY910 pKa = 9.26FVKK913 pKa = 10.79LNMQGKK919 pKa = 9.23EE920 pKa = 3.89IVKK923 pKa = 10.6KK924 pKa = 10.71LVLNNKK930 pKa = 8.99
Molecular weight: 98.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A399CY40|A0A399CY40_9BACT Response regulator OS=Mariniphaga sediminis OX=1628158 GN=D1164_19880 PE=4 SV=1
MM1 pKa = 7.3VFYY4 pKa = 10.88SCQYY8 pKa = 10.16TNYY11 pKa = 9.95CFANQPYY18 pKa = 9.58KK19 pKa = 10.93LSIFWGNACMLLYY32 pKa = 10.22ILEE35 pKa = 4.41GNIFFPNLLTRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.93TLFSDD54 pKa = 3.58AKK56 pKa = 10.05VRR58 pKa = 11.84LHH60 pKa = 6.13LRR62 pKa = 11.84PVTSRR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 8.36TGWHH73 pKa = 5.91NNWVTGSKK81 pKa = 9.22TEE83 pKa = 4.1NVSRR87 pKa = 11.84RR88 pKa = 11.84QKK90 pKa = 9.92KK91 pKa = 9.15CRR93 pKa = 11.84EE94 pKa = 3.84IILRR98 pKa = 3.99
MM1 pKa = 7.3VFYY4 pKa = 10.88SCQYY8 pKa = 10.16TNYY11 pKa = 9.95CFANQPYY18 pKa = 9.58KK19 pKa = 10.93LSIFWGNACMLLYY32 pKa = 10.22ILEE35 pKa = 4.41GNIFFPNLLTRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 4.93TLFSDD54 pKa = 3.58AKK56 pKa = 10.05VRR58 pKa = 11.84LHH60 pKa = 6.13LRR62 pKa = 11.84PVTSRR67 pKa = 11.84RR68 pKa = 11.84KK69 pKa = 8.36TGWHH73 pKa = 5.91NNWVTGSKK81 pKa = 9.22TEE83 pKa = 4.1NVSRR87 pKa = 11.84RR88 pKa = 11.84QKK90 pKa = 9.92KK91 pKa = 9.15CRR93 pKa = 11.84EE94 pKa = 3.84IILRR98 pKa = 3.99
Molecular weight: 11.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1693228 |
28 |
3263 |
374.9 |
42.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.409 ± 0.035 | 0.865 ± 0.015 |
5.275 ± 0.026 | 7.031 ± 0.035 |
5.262 ± 0.026 | 7.109 ± 0.037 |
1.88 ± 0.016 | 7.147 ± 0.031 |
6.883 ± 0.038 | 9.112 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.352 ± 0.015 | 5.525 ± 0.027 |
3.963 ± 0.019 | 3.337 ± 0.021 |
4.24 ± 0.025 | 6.336 ± 0.029 |
5.331 ± 0.04 | 6.486 ± 0.024 |
1.413 ± 0.015 | 4.044 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
