
Bacillus sp. AFS037270
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
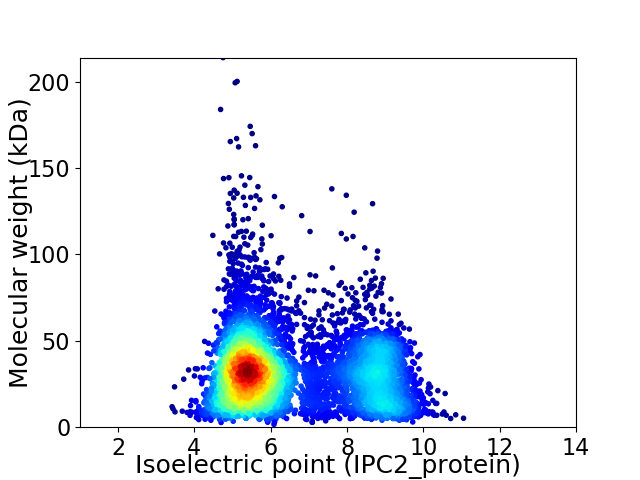
Virtual 2D-PAGE plot for 5980 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C1YUP9|A0A2C1YUP9_9BACI Uncharacterized protein OS=Bacillus sp. AFS037270 OX=2033499 GN=COD92_25430 PE=4 SV=1
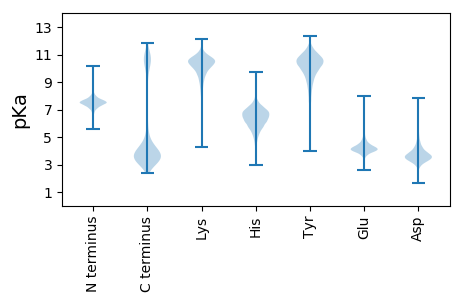
MM1 pKa = 7.58FNRR4 pKa = 11.84IDD6 pKa = 3.48KK7 pKa = 10.04EE8 pKa = 4.31LYY10 pKa = 9.05GNEE13 pKa = 4.0YY14 pKa = 10.49QDD16 pKa = 4.45ANLNGVRR23 pKa = 11.84QEE25 pKa = 3.88MGLPSGAEE33 pKa = 3.87AGQQAMDD40 pKa = 3.95HH41 pKa = 6.69GIDD44 pKa = 3.27NDD46 pKa = 3.82LYY48 pKa = 9.24EE49 pKa = 4.85NEE51 pKa = 4.65YY52 pKa = 10.82EE53 pKa = 5.21DD54 pKa = 5.47SDD56 pKa = 4.45FNDD59 pKa = 2.97VRR61 pKa = 11.84QQMGLPNGAEE71 pKa = 4.04AGQQAVQYY79 pKa = 10.82GIDD82 pKa = 3.39NNLYY86 pKa = 8.35EE87 pKa = 5.03NEE89 pKa = 4.16YY90 pKa = 10.17KK91 pKa = 10.75DD92 pKa = 4.35SGFSDD97 pKa = 3.56VRR99 pKa = 11.84QQMVLPTRR107 pKa = 11.84ADD109 pKa = 3.46AEE111 pKa = 4.32QQAMHH116 pKa = 6.72HH117 pKa = 6.54GIDD120 pKa = 3.29NDD122 pKa = 4.09LYY124 pKa = 9.85GNEE127 pKa = 4.44YY128 pKa = 10.34EE129 pKa = 5.9DD130 pKa = 5.04SDD132 pKa = 4.5FNDD135 pKa = 2.97VRR137 pKa = 11.84QQMGLPNGAEE147 pKa = 4.27VEE149 pKa = 4.34QQAGQDD155 pKa = 4.04GIDD158 pKa = 3.19NCLYY162 pKa = 9.17EE163 pKa = 5.32NEE165 pKa = 4.25YY166 pKa = 11.04EE167 pKa = 5.21DD168 pKa = 4.69YY169 pKa = 11.28DD170 pKa = 4.46FNSVRR175 pKa = 11.84QQMGLPSGAEE185 pKa = 3.85TGQQAMHH192 pKa = 5.82QAGYY196 pKa = 9.7QAGFQAGFQAGFQTGFQTGFQAGYY220 pKa = 7.76QTGNQTGLQTGAQAGVPTGTQVGSQTGAQMGTQDD254 pKa = 3.44GAQAGTQTEE263 pKa = 4.59TQDD266 pKa = 3.05GTQAGTQTGAQDD278 pKa = 3.77GAQVEE283 pKa = 4.52AQTEE287 pKa = 4.28QQHH290 pKa = 5.48PHH292 pKa = 6.88AGHH295 pKa = 5.84QFGPRR300 pKa = 11.84DD301 pKa = 3.53LSYY304 pKa = 10.92EE305 pKa = 3.97
MM1 pKa = 7.58FNRR4 pKa = 11.84IDD6 pKa = 3.48KK7 pKa = 10.04EE8 pKa = 4.31LYY10 pKa = 9.05GNEE13 pKa = 4.0YY14 pKa = 10.49QDD16 pKa = 4.45ANLNGVRR23 pKa = 11.84QEE25 pKa = 3.88MGLPSGAEE33 pKa = 3.87AGQQAMDD40 pKa = 3.95HH41 pKa = 6.69GIDD44 pKa = 3.27NDD46 pKa = 3.82LYY48 pKa = 9.24EE49 pKa = 4.85NEE51 pKa = 4.65YY52 pKa = 10.82EE53 pKa = 5.21DD54 pKa = 5.47SDD56 pKa = 4.45FNDD59 pKa = 2.97VRR61 pKa = 11.84QQMGLPNGAEE71 pKa = 4.04AGQQAVQYY79 pKa = 10.82GIDD82 pKa = 3.39NNLYY86 pKa = 8.35EE87 pKa = 5.03NEE89 pKa = 4.16YY90 pKa = 10.17KK91 pKa = 10.75DD92 pKa = 4.35SGFSDD97 pKa = 3.56VRR99 pKa = 11.84QQMVLPTRR107 pKa = 11.84ADD109 pKa = 3.46AEE111 pKa = 4.32QQAMHH116 pKa = 6.72HH117 pKa = 6.54GIDD120 pKa = 3.29NDD122 pKa = 4.09LYY124 pKa = 9.85GNEE127 pKa = 4.44YY128 pKa = 10.34EE129 pKa = 5.9DD130 pKa = 5.04SDD132 pKa = 4.5FNDD135 pKa = 2.97VRR137 pKa = 11.84QQMGLPNGAEE147 pKa = 4.27VEE149 pKa = 4.34QQAGQDD155 pKa = 4.04GIDD158 pKa = 3.19NCLYY162 pKa = 9.17EE163 pKa = 5.32NEE165 pKa = 4.25YY166 pKa = 11.04EE167 pKa = 5.21DD168 pKa = 4.69YY169 pKa = 11.28DD170 pKa = 4.46FNSVRR175 pKa = 11.84QQMGLPSGAEE185 pKa = 3.85TGQQAMHH192 pKa = 5.82QAGYY196 pKa = 9.7QAGFQAGFQAGFQTGFQTGFQAGYY220 pKa = 7.76QTGNQTGLQTGAQAGVPTGTQVGSQTGAQMGTQDD254 pKa = 3.44GAQAGTQTEE263 pKa = 4.59TQDD266 pKa = 3.05GTQAGTQTGAQDD278 pKa = 3.77GAQVEE283 pKa = 4.52AQTEE287 pKa = 4.28QQHH290 pKa = 5.48PHH292 pKa = 6.88AGHH295 pKa = 5.84QFGPRR300 pKa = 11.84DD301 pKa = 3.53LSYY304 pKa = 10.92EE305 pKa = 3.97
Molecular weight: 33.15 kDa
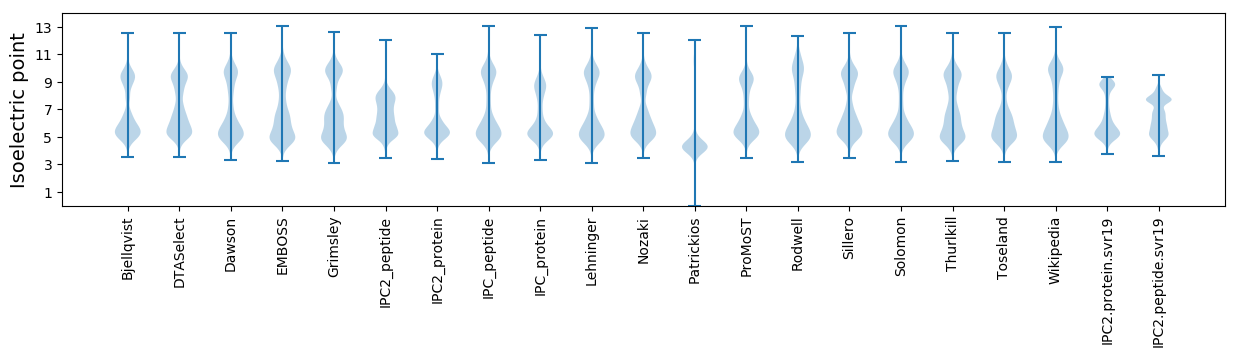
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C1YCP8|A0A2C1YCP8_9BACI Thiol-disulfide oxidoreductase OS=Bacillus sp. AFS037270 OX=2033499 GN=COD92_19595 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.24QPNKK9 pKa = 8.16RR10 pKa = 11.84KK11 pKa = 9.54HH12 pKa = 5.99SKK14 pKa = 8.79VHH16 pKa = 5.85GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.7VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.24QPNKK9 pKa = 8.16RR10 pKa = 11.84KK11 pKa = 9.54HH12 pKa = 5.99SKK14 pKa = 8.79VHH16 pKa = 5.85GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.7VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1805640 |
15 |
2017 |
301.9 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.931 ± 0.031 | 0.785 ± 0.008 |
4.852 ± 0.021 | 7.009 ± 0.039 |
4.696 ± 0.023 | 7.114 ± 0.028 |
2.063 ± 0.015 | 7.984 ± 0.033 |
6.972 ± 0.025 | 9.853 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.692 ± 0.015 | 4.64 ± 0.019 |
3.743 ± 0.019 | 3.646 ± 0.019 |
3.884 ± 0.021 | 6.069 ± 0.022 |
5.433 ± 0.023 | 6.96 ± 0.026 |
1.127 ± 0.013 | 3.546 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
