
Aureitalea sp. RR4-38
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Pukyongia
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
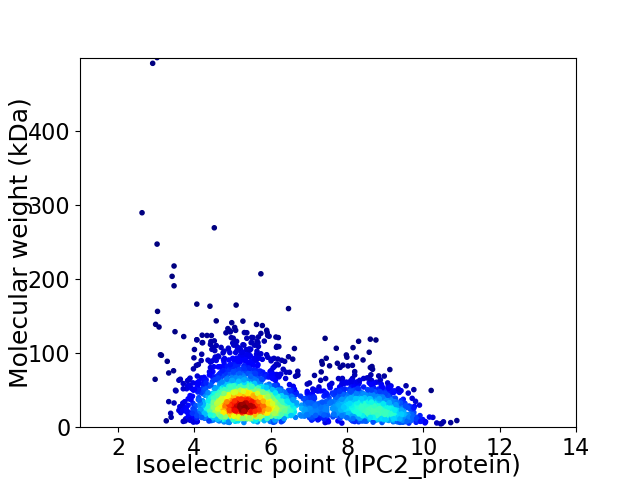
Virtual 2D-PAGE plot for 2826 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S0HXP2|A0A2S0HXP2_9FLAO Uncharacterized protein OS=Aureitalea sp. RR4-38 OX=2094025 GN=C5O00_09530 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.26ILIPSVLQVVLLFNFALIYY21 pKa = 10.51GQAQFEE27 pKa = 4.34NQAEE31 pKa = 4.3SLGAGIAYY39 pKa = 10.29GNIQLGAGVSFFDD52 pKa = 4.32YY53 pKa = 11.33NNDD56 pKa = 3.41GLDD59 pKa = 4.84DD60 pKa = 3.67ITIPASNSRR69 pKa = 11.84NFTFLKK75 pKa = 10.91NMGGTFQEE83 pKa = 4.4EE84 pKa = 4.16NLGIDD89 pKa = 3.4SNGFQSRR96 pKa = 11.84QAMWVDD102 pKa = 4.15FDD104 pKa = 4.93NDD106 pKa = 3.21GDD108 pKa = 3.93NDD110 pKa = 3.82FFATSDD116 pKa = 3.68QDD118 pKa = 3.24WSRR121 pKa = 11.84LYY123 pKa = 10.81QNNGDD128 pKa = 3.9GTFTDD133 pKa = 3.16ITLAAGLPTVIYY145 pKa = 8.37DD146 pKa = 3.81TYY148 pKa = 10.11GAAWGDD154 pKa = 4.01FNNDD158 pKa = 2.18RR159 pKa = 11.84HH160 pKa = 7.45LDD162 pKa = 3.5VFYY165 pKa = 10.87SIRR168 pKa = 11.84DD169 pKa = 3.55EE170 pKa = 4.1NQIIPNILYY179 pKa = 10.31QNNGDD184 pKa = 3.78GTFTNVTIAAGLEE197 pKa = 4.14TTGYY201 pKa = 11.15LSFCASFFDD210 pKa = 3.8MDD212 pKa = 4.36KK213 pKa = 11.5DD214 pKa = 3.82GDD216 pKa = 3.61QDD218 pKa = 5.11LYY220 pKa = 11.14VANDD224 pKa = 3.93KK225 pKa = 11.05YY226 pKa = 11.4DD227 pKa = 3.71SPNLMYY233 pKa = 10.67RR234 pKa = 11.84NNGDD238 pKa = 3.44GTFTNVSASSGTGIEE253 pKa = 3.94IDD255 pKa = 4.26AMSTTIGDD263 pKa = 3.88YY264 pKa = 11.56NNDD267 pKa = 3.25GWLDD271 pKa = 3.34IYY273 pKa = 9.74VTNTEE278 pKa = 4.37FTPPPPGVNGNVLFRR293 pKa = 11.84NNADD297 pKa = 3.39GTFTNVAASTGTIFNSVGWGAVFLDD322 pKa = 4.02GDD324 pKa = 3.96NEE326 pKa = 4.46GFTDD330 pKa = 4.96LYY332 pKa = 11.04VSGEE336 pKa = 4.17FEE338 pKa = 4.18VDD340 pKa = 3.31PVYY343 pKa = 10.83LPSAYY348 pKa = 10.4YY349 pKa = 10.22EE350 pKa = 4.11NQGDD354 pKa = 4.0HH355 pKa = 6.22TFVIPTGIGFEE366 pKa = 3.99NDD368 pKa = 2.76TRR370 pKa = 11.84QSYY373 pKa = 10.96GNAIGDD379 pKa = 4.04IDD381 pKa = 4.27NDD383 pKa = 4.45GYY385 pKa = 10.47PDD387 pKa = 3.25IVVMNNNDD395 pKa = 3.26QNIFLWKK402 pKa = 8.38NTSIQNHH409 pKa = 5.04NWLKK413 pKa = 10.86VKK415 pKa = 11.08LEE417 pKa = 3.99GTQGNRR423 pKa = 11.84MGIGSFIEE431 pKa = 4.01VGTTHH436 pKa = 6.73GTQLQFTVCGEE447 pKa = 4.37GYY449 pKa = 10.46LGQNSAYY456 pKa = 10.11EE457 pKa = 4.1FFGLNDD463 pKa = 3.41ATNVDD468 pKa = 4.85FVRR471 pKa = 11.84VTWPSGIVDD480 pKa = 3.76YY481 pKa = 11.46FEE483 pKa = 5.25DD484 pKa = 4.37LNVNQHH490 pKa = 4.36ITIIEE495 pKa = 4.19GTGILSSDD503 pKa = 3.57SASFAVFSMYY513 pKa = 10.41PNPASEE519 pKa = 4.77LVFLNFPSEE528 pKa = 4.25AEE530 pKa = 4.04RR531 pKa = 11.84QLVVLDD537 pKa = 3.52VTGKK541 pKa = 9.93VVISEE546 pKa = 4.45EE547 pKa = 4.05IFGNTHH553 pKa = 6.05QMDD556 pKa = 4.49LSVISGGVYY565 pKa = 9.11IVEE568 pKa = 3.98VRR570 pKa = 11.84FNGNVHH576 pKa = 6.09RR577 pKa = 11.84KK578 pKa = 9.71KK579 pKa = 10.65LIKK582 pKa = 10.15HH583 pKa = 5.84
MM1 pKa = 7.76KK2 pKa = 10.26ILIPSVLQVVLLFNFALIYY21 pKa = 10.51GQAQFEE27 pKa = 4.34NQAEE31 pKa = 4.3SLGAGIAYY39 pKa = 10.29GNIQLGAGVSFFDD52 pKa = 4.32YY53 pKa = 11.33NNDD56 pKa = 3.41GLDD59 pKa = 4.84DD60 pKa = 3.67ITIPASNSRR69 pKa = 11.84NFTFLKK75 pKa = 10.91NMGGTFQEE83 pKa = 4.4EE84 pKa = 4.16NLGIDD89 pKa = 3.4SNGFQSRR96 pKa = 11.84QAMWVDD102 pKa = 4.15FDD104 pKa = 4.93NDD106 pKa = 3.21GDD108 pKa = 3.93NDD110 pKa = 3.82FFATSDD116 pKa = 3.68QDD118 pKa = 3.24WSRR121 pKa = 11.84LYY123 pKa = 10.81QNNGDD128 pKa = 3.9GTFTDD133 pKa = 3.16ITLAAGLPTVIYY145 pKa = 8.37DD146 pKa = 3.81TYY148 pKa = 10.11GAAWGDD154 pKa = 4.01FNNDD158 pKa = 2.18RR159 pKa = 11.84HH160 pKa = 7.45LDD162 pKa = 3.5VFYY165 pKa = 10.87SIRR168 pKa = 11.84DD169 pKa = 3.55EE170 pKa = 4.1NQIIPNILYY179 pKa = 10.31QNNGDD184 pKa = 3.78GTFTNVTIAAGLEE197 pKa = 4.14TTGYY201 pKa = 11.15LSFCASFFDD210 pKa = 3.8MDD212 pKa = 4.36KK213 pKa = 11.5DD214 pKa = 3.82GDD216 pKa = 3.61QDD218 pKa = 5.11LYY220 pKa = 11.14VANDD224 pKa = 3.93KK225 pKa = 11.05YY226 pKa = 11.4DD227 pKa = 3.71SPNLMYY233 pKa = 10.67RR234 pKa = 11.84NNGDD238 pKa = 3.44GTFTNVSASSGTGIEE253 pKa = 3.94IDD255 pKa = 4.26AMSTTIGDD263 pKa = 3.88YY264 pKa = 11.56NNDD267 pKa = 3.25GWLDD271 pKa = 3.34IYY273 pKa = 9.74VTNTEE278 pKa = 4.37FTPPPPGVNGNVLFRR293 pKa = 11.84NNADD297 pKa = 3.39GTFTNVAASTGTIFNSVGWGAVFLDD322 pKa = 4.02GDD324 pKa = 3.96NEE326 pKa = 4.46GFTDD330 pKa = 4.96LYY332 pKa = 11.04VSGEE336 pKa = 4.17FEE338 pKa = 4.18VDD340 pKa = 3.31PVYY343 pKa = 10.83LPSAYY348 pKa = 10.4YY349 pKa = 10.22EE350 pKa = 4.11NQGDD354 pKa = 4.0HH355 pKa = 6.22TFVIPTGIGFEE366 pKa = 3.99NDD368 pKa = 2.76TRR370 pKa = 11.84QSYY373 pKa = 10.96GNAIGDD379 pKa = 4.04IDD381 pKa = 4.27NDD383 pKa = 4.45GYY385 pKa = 10.47PDD387 pKa = 3.25IVVMNNNDD395 pKa = 3.26QNIFLWKK402 pKa = 8.38NTSIQNHH409 pKa = 5.04NWLKK413 pKa = 10.86VKK415 pKa = 11.08LEE417 pKa = 3.99GTQGNRR423 pKa = 11.84MGIGSFIEE431 pKa = 4.01VGTTHH436 pKa = 6.73GTQLQFTVCGEE447 pKa = 4.37GYY449 pKa = 10.46LGQNSAYY456 pKa = 10.11EE457 pKa = 4.1FFGLNDD463 pKa = 3.41ATNVDD468 pKa = 4.85FVRR471 pKa = 11.84VTWPSGIVDD480 pKa = 3.76YY481 pKa = 11.46FEE483 pKa = 5.25DD484 pKa = 4.37LNVNQHH490 pKa = 4.36ITIIEE495 pKa = 4.19GTGILSSDD503 pKa = 3.57SASFAVFSMYY513 pKa = 10.41PNPASEE519 pKa = 4.77LVFLNFPSEE528 pKa = 4.25AEE530 pKa = 4.04RR531 pKa = 11.84QLVVLDD537 pKa = 3.52VTGKK541 pKa = 9.93VVISEE546 pKa = 4.45EE547 pKa = 4.05IFGNTHH553 pKa = 6.05QMDD556 pKa = 4.49LSVISGGVYY565 pKa = 9.11IVEE568 pKa = 3.98VRR570 pKa = 11.84FNGNVHH576 pKa = 6.09RR577 pKa = 11.84KK578 pKa = 9.71KK579 pKa = 10.65LIKK582 pKa = 10.15HH583 pKa = 5.84
Molecular weight: 64.21 kDa
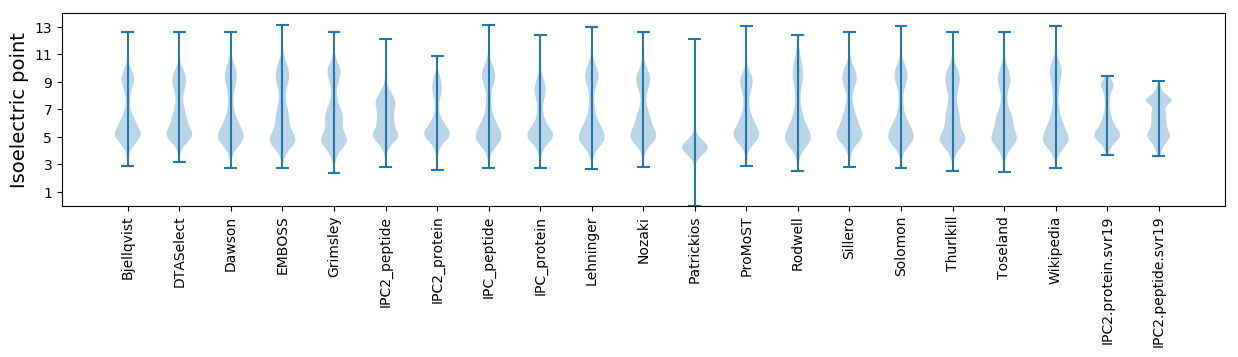
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S0HTP0|A0A2S0HTP0_9FLAO DNA gyrase subunit B OS=Aureitalea sp. RR4-38 OX=2094025 GN=gyrB PE=3 SV=1
MM1 pKa = 7.46SFNSASFKK9 pKa = 10.55AASLRR14 pKa = 11.84ANSFIAASFSAASFIAASFNAASFNAASFKK44 pKa = 10.65AASLRR49 pKa = 11.84ANSFNAASFRR59 pKa = 11.84AASLRR64 pKa = 11.84AASFMAASFKK74 pKa = 10.78AASFKK79 pKa = 10.77AASLRR84 pKa = 11.84AASFF88 pKa = 3.37
MM1 pKa = 7.46SFNSASFKK9 pKa = 10.55AASLRR14 pKa = 11.84ANSFIAASFSAASFIAASFNAASFNAASFKK44 pKa = 10.65AASLRR49 pKa = 11.84ANSFNAASFRR59 pKa = 11.84AASLRR64 pKa = 11.84AASFMAASFKK74 pKa = 10.78AASFKK79 pKa = 10.77AASLRR84 pKa = 11.84AASFF88 pKa = 3.37
Molecular weight: 8.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
970976 |
38 |
4750 |
343.6 |
38.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.757 ± 0.041 | 0.762 ± 0.017 |
5.857 ± 0.057 | 6.83 ± 0.053 |
5.045 ± 0.037 | 6.828 ± 0.052 |
1.79 ± 0.025 | 7.438 ± 0.039 |
6.686 ± 0.076 | 9.28 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.024 | 5.63 ± 0.044 |
3.687 ± 0.035 | 3.264 ± 0.024 |
4.003 ± 0.042 | 6.401 ± 0.035 |
5.842 ± 0.073 | 6.493 ± 0.043 |
1.075 ± 0.017 | 4.002 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
