
Apis mellifera associated microvirus 27
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
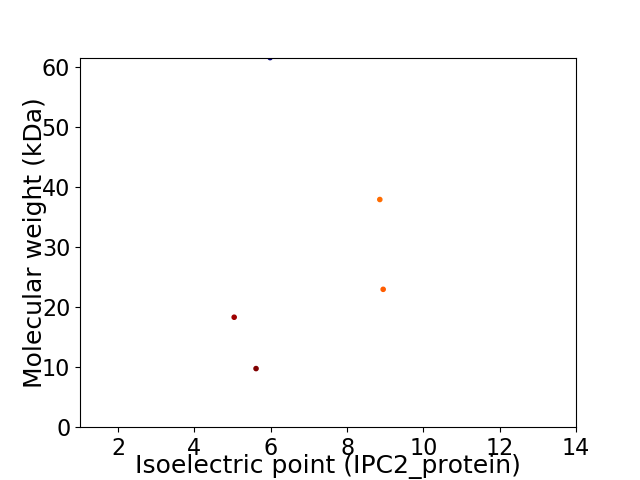
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3Q8U5B7|A0A3Q8U5B7_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 27 OX=2494755 PE=4 SV=1
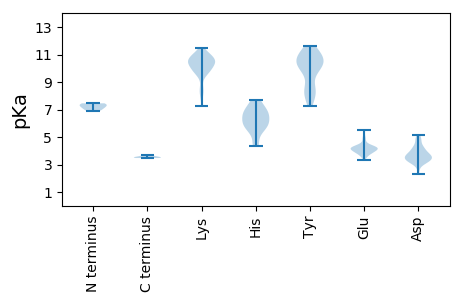
MM1 pKa = 7.45KK2 pKa = 10.36KK3 pKa = 8.41ITEE6 pKa = 3.91EE7 pKa = 4.62SYY9 pKa = 10.87VEE11 pKa = 4.08TYY13 pKa = 8.01TRR15 pKa = 11.84KK16 pKa = 10.4NGTFGYY22 pKa = 8.65RR23 pKa = 11.84TINNDD28 pKa = 2.34VSMADD33 pKa = 3.29PTGVEE38 pKa = 4.0EE39 pKa = 5.47AEE41 pKa = 4.2ITSIMSNFAKK51 pKa = 10.04TGRR54 pKa = 11.84LPLRR58 pKa = 11.84GGQPGLYY65 pKa = 10.14SPDD68 pKa = 3.07EE69 pKa = 4.09VVPNGKK75 pKa = 9.42DD76 pKa = 3.15LLSAVTLVQEE86 pKa = 3.78ATEE89 pKa = 4.24RR90 pKa = 11.84FEE92 pKa = 4.39QLPSTLRR99 pKa = 11.84EE100 pKa = 3.82RR101 pKa = 11.84FGNDD105 pKa = 3.76PIKK108 pKa = 10.58MYY110 pKa = 11.09DD111 pKa = 3.62FLSDD115 pKa = 3.73PKK117 pKa = 11.33NMPEE121 pKa = 3.74AVKK124 pKa = 10.59LGIIQQPVSKK134 pKa = 10.65NDD136 pKa = 3.45EE137 pKa = 4.31LNDD140 pKa = 5.09DD141 pKa = 4.33IEE143 pKa = 4.71TKK145 pKa = 8.27NTRR148 pKa = 11.84TTKK151 pKa = 7.27TQKK154 pKa = 10.72AKK156 pKa = 10.71EE157 pKa = 4.03KK158 pKa = 9.75TPEE161 pKa = 4.05PEE163 pKa = 3.57
MM1 pKa = 7.45KK2 pKa = 10.36KK3 pKa = 8.41ITEE6 pKa = 3.91EE7 pKa = 4.62SYY9 pKa = 10.87VEE11 pKa = 4.08TYY13 pKa = 8.01TRR15 pKa = 11.84KK16 pKa = 10.4NGTFGYY22 pKa = 8.65RR23 pKa = 11.84TINNDD28 pKa = 2.34VSMADD33 pKa = 3.29PTGVEE38 pKa = 4.0EE39 pKa = 5.47AEE41 pKa = 4.2ITSIMSNFAKK51 pKa = 10.04TGRR54 pKa = 11.84LPLRR58 pKa = 11.84GGQPGLYY65 pKa = 10.14SPDD68 pKa = 3.07EE69 pKa = 4.09VVPNGKK75 pKa = 9.42DD76 pKa = 3.15LLSAVTLVQEE86 pKa = 3.78ATEE89 pKa = 4.24RR90 pKa = 11.84FEE92 pKa = 4.39QLPSTLRR99 pKa = 11.84EE100 pKa = 3.82RR101 pKa = 11.84FGNDD105 pKa = 3.76PIKK108 pKa = 10.58MYY110 pKa = 11.09DD111 pKa = 3.62FLSDD115 pKa = 3.73PKK117 pKa = 11.33NMPEE121 pKa = 3.74AVKK124 pKa = 10.59LGIIQQPVSKK134 pKa = 10.65NDD136 pKa = 3.45EE137 pKa = 4.31LNDD140 pKa = 5.09DD141 pKa = 4.33IEE143 pKa = 4.71TKK145 pKa = 8.27NTRR148 pKa = 11.84TTKK151 pKa = 7.27TQKK154 pKa = 10.72AKK156 pKa = 10.71EE157 pKa = 4.03KK158 pKa = 9.75TPEE161 pKa = 4.05PEE163 pKa = 3.57
Molecular weight: 18.3 kDa
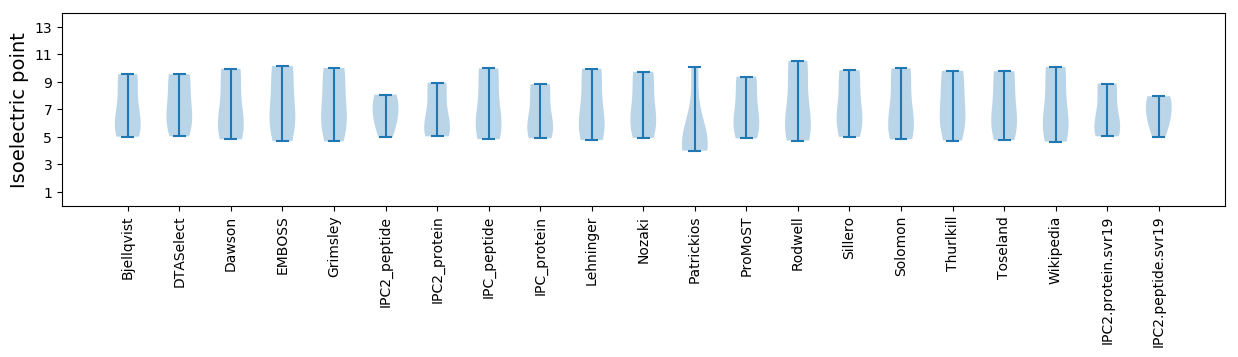
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTQ5|A0A3S8UTQ5_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 27 OX=2494755 PE=3 SV=1
MM1 pKa = 7.25QCINKK6 pKa = 9.4IKK8 pKa = 10.73AGFTRR13 pKa = 11.84TGDD16 pKa = 3.44LTFKK20 pKa = 10.35PRR22 pKa = 11.84QADD25 pKa = 3.36PGIVGFEE32 pKa = 4.11LEE34 pKa = 4.49CRR36 pKa = 11.84KK37 pKa = 10.02CLPCRR42 pKa = 11.84LNIARR47 pKa = 11.84EE48 pKa = 3.7KK49 pKa = 10.58AIRR52 pKa = 11.84AVHH55 pKa = 5.75EE56 pKa = 4.36AEE58 pKa = 4.1CHH60 pKa = 5.48KK61 pKa = 11.16KK62 pKa = 10.67NIFLTLTYY70 pKa = 10.83APGKK74 pKa = 10.05LKK76 pKa = 10.61SPRR79 pKa = 11.84LNYY82 pKa = 10.79GDD84 pKa = 4.06FQLFMKK90 pKa = 10.54RR91 pKa = 11.84LRR93 pKa = 11.84EE94 pKa = 3.81LHH96 pKa = 6.0TRR98 pKa = 11.84DD99 pKa = 4.68VLDD102 pKa = 4.77PDD104 pKa = 4.24LKK106 pKa = 11.09KK107 pKa = 10.95EE108 pKa = 3.87MSISYY113 pKa = 8.46MVTGEE118 pKa = 4.11YY119 pKa = 10.62GDD121 pKa = 3.75KK122 pKa = 10.58NKK124 pKa = 10.21RR125 pKa = 11.84PHH127 pKa = 4.33WHH129 pKa = 7.1AIIFNYY135 pKa = 10.14SPPDD139 pKa = 3.61LMLHH143 pKa = 5.82RR144 pKa = 11.84TDD146 pKa = 3.49QNDD149 pKa = 3.41YY150 pKa = 11.29QIFKK154 pKa = 10.94SKK156 pKa = 10.4IIDD159 pKa = 3.85EE160 pKa = 4.28LWGFNDD166 pKa = 5.12PEE168 pKa = 4.26NCPNEE173 pKa = 4.23IGSVTIEE180 pKa = 3.96SAGYY184 pKa = 7.27VARR187 pKa = 11.84YY188 pKa = 7.94AAKK191 pKa = 10.45KK192 pKa = 8.19LTHH195 pKa = 6.23GQDD198 pKa = 3.33QEE200 pKa = 4.9HH201 pKa = 7.38DD202 pKa = 3.58FHH204 pKa = 6.83PVHH207 pKa = 6.26KK208 pKa = 8.72TSCVNAVGKK217 pKa = 10.3RR218 pKa = 11.84WIEE221 pKa = 3.52KK222 pKa = 9.94HH223 pKa = 4.34YY224 pKa = 10.69QQVFKK229 pKa = 10.87LGYY232 pKa = 8.32VWLPNGGRR240 pKa = 11.84TKK242 pKa = 10.31IPRR245 pKa = 11.84YY246 pKa = 9.27YY247 pKa = 11.03VDD249 pKa = 3.3WLKK252 pKa = 11.18KK253 pKa = 9.72NHH255 pKa = 6.59PEE257 pKa = 3.49EE258 pKa = 4.32HH259 pKa = 6.53KK260 pKa = 11.23NYY262 pKa = 7.56VTKK265 pKa = 10.37IRR267 pKa = 11.84HH268 pKa = 5.71KK269 pKa = 10.19LQLAAAEE276 pKa = 4.16KK277 pKa = 10.54SRR279 pKa = 11.84NEE281 pKa = 3.9EE282 pKa = 3.56IQYY285 pKa = 8.69LTEE288 pKa = 4.98LMNLPHH294 pKa = 7.64GYY296 pKa = 9.8PMPEE300 pKa = 4.26SKK302 pKa = 10.67KK303 pKa = 10.8KK304 pKa = 9.93MSLTILKK311 pKa = 10.16QKK313 pKa = 10.18FKK315 pKa = 11.11KK316 pKa = 10.38LQEE319 pKa = 4.07KK320 pKa = 10.63LKK322 pKa = 10.97LL323 pKa = 3.7
MM1 pKa = 7.25QCINKK6 pKa = 9.4IKK8 pKa = 10.73AGFTRR13 pKa = 11.84TGDD16 pKa = 3.44LTFKK20 pKa = 10.35PRR22 pKa = 11.84QADD25 pKa = 3.36PGIVGFEE32 pKa = 4.11LEE34 pKa = 4.49CRR36 pKa = 11.84KK37 pKa = 10.02CLPCRR42 pKa = 11.84LNIARR47 pKa = 11.84EE48 pKa = 3.7KK49 pKa = 10.58AIRR52 pKa = 11.84AVHH55 pKa = 5.75EE56 pKa = 4.36AEE58 pKa = 4.1CHH60 pKa = 5.48KK61 pKa = 11.16KK62 pKa = 10.67NIFLTLTYY70 pKa = 10.83APGKK74 pKa = 10.05LKK76 pKa = 10.61SPRR79 pKa = 11.84LNYY82 pKa = 10.79GDD84 pKa = 4.06FQLFMKK90 pKa = 10.54RR91 pKa = 11.84LRR93 pKa = 11.84EE94 pKa = 3.81LHH96 pKa = 6.0TRR98 pKa = 11.84DD99 pKa = 4.68VLDD102 pKa = 4.77PDD104 pKa = 4.24LKK106 pKa = 11.09KK107 pKa = 10.95EE108 pKa = 3.87MSISYY113 pKa = 8.46MVTGEE118 pKa = 4.11YY119 pKa = 10.62GDD121 pKa = 3.75KK122 pKa = 10.58NKK124 pKa = 10.21RR125 pKa = 11.84PHH127 pKa = 4.33WHH129 pKa = 7.1AIIFNYY135 pKa = 10.14SPPDD139 pKa = 3.61LMLHH143 pKa = 5.82RR144 pKa = 11.84TDD146 pKa = 3.49QNDD149 pKa = 3.41YY150 pKa = 11.29QIFKK154 pKa = 10.94SKK156 pKa = 10.4IIDD159 pKa = 3.85EE160 pKa = 4.28LWGFNDD166 pKa = 5.12PEE168 pKa = 4.26NCPNEE173 pKa = 4.23IGSVTIEE180 pKa = 3.96SAGYY184 pKa = 7.27VARR187 pKa = 11.84YY188 pKa = 7.94AAKK191 pKa = 10.45KK192 pKa = 8.19LTHH195 pKa = 6.23GQDD198 pKa = 3.33QEE200 pKa = 4.9HH201 pKa = 7.38DD202 pKa = 3.58FHH204 pKa = 6.83PVHH207 pKa = 6.26KK208 pKa = 8.72TSCVNAVGKK217 pKa = 10.3RR218 pKa = 11.84WIEE221 pKa = 3.52KK222 pKa = 9.94HH223 pKa = 4.34YY224 pKa = 10.69QQVFKK229 pKa = 10.87LGYY232 pKa = 8.32VWLPNGGRR240 pKa = 11.84TKK242 pKa = 10.31IPRR245 pKa = 11.84YY246 pKa = 9.27YY247 pKa = 11.03VDD249 pKa = 3.3WLKK252 pKa = 11.18KK253 pKa = 9.72NHH255 pKa = 6.59PEE257 pKa = 3.49EE258 pKa = 4.32HH259 pKa = 6.53KK260 pKa = 11.23NYY262 pKa = 7.56VTKK265 pKa = 10.37IRR267 pKa = 11.84HH268 pKa = 5.71KK269 pKa = 10.19LQLAAAEE276 pKa = 4.16KK277 pKa = 10.54SRR279 pKa = 11.84NEE281 pKa = 3.9EE282 pKa = 3.56IQYY285 pKa = 8.69LTEE288 pKa = 4.98LMNLPHH294 pKa = 7.64GYY296 pKa = 9.8PMPEE300 pKa = 4.26SKK302 pKa = 10.67KK303 pKa = 10.8KK304 pKa = 9.93MSLTILKK311 pKa = 10.16QKK313 pKa = 10.18FKK315 pKa = 11.11KK316 pKa = 10.38LQEE319 pKa = 4.07KK320 pKa = 10.63LKK322 pKa = 10.97LL323 pKa = 3.7
Molecular weight: 37.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1334 |
84 |
545 |
266.8 |
30.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.946 ± 2.338 | 0.675 ± 0.487 |
4.873 ± 0.74 | 6.072 ± 1.073 |
5.247 ± 1.437 | 6.372 ± 0.513 |
2.324 ± 0.815 | 4.648 ± 0.467 |
7.271 ± 2.1 | 7.946 ± 0.638 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.544 | 5.322 ± 0.275 |
6.297 ± 0.748 | 5.172 ± 0.618 |
4.873 ± 0.711 | 5.922 ± 0.949 |
6.897 ± 0.894 | 4.723 ± 0.617 |
0.975 ± 0.366 | 3.973 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
