
Clostridium sp. MD294
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
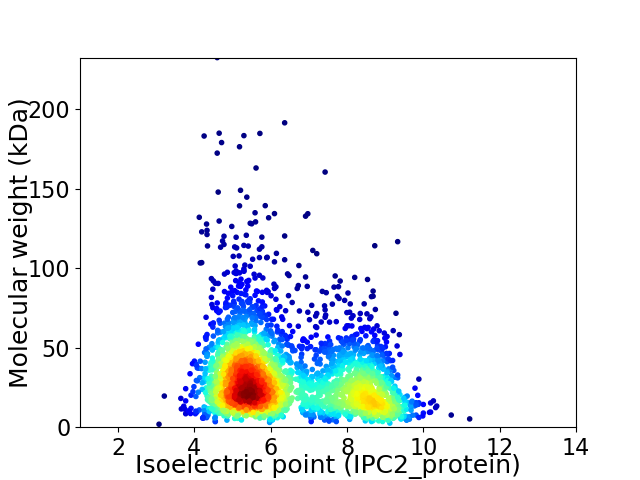
Virtual 2D-PAGE plot for 2792 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|N1ZF63|N1ZF63_9CLOT Uncharacterized protein OS=Clostridium sp. MD294 OX=97138 GN=C820_01881 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 10.42KK3 pKa = 9.98RR4 pKa = 11.84VLGLLLCSVMAFSMFAAGCGGSNNAPAPEE33 pKa = 3.91QTEE36 pKa = 4.15NGQNTSTSEE45 pKa = 4.03GGLKK49 pKa = 9.76IAIVSSPSGVDD60 pKa = 3.39DD61 pKa = 5.34GSFNQNNYY69 pKa = 10.33EE70 pKa = 4.37GIQSFIANHH79 pKa = 6.99PDD81 pKa = 3.21ASVTPVKK88 pKa = 10.6EE89 pKa = 4.07EE90 pKa = 4.16SGDD93 pKa = 3.34TAAAVQAVADD103 pKa = 3.93IVADD107 pKa = 3.66YY108 pKa = 11.18NVIVCCGFQFAGIGAITQDD127 pKa = 3.49NPNVNFILVDD137 pKa = 4.03SNPTDD142 pKa = 3.46AQGQEE147 pKa = 3.88INTEE151 pKa = 3.98NIYY154 pKa = 11.54AMTFAEE160 pKa = 4.3QEE162 pKa = 4.14SGFYY166 pKa = 10.56AGVAAALEE174 pKa = 4.35TQSNKK179 pKa = 9.18VAVVNGIAYY188 pKa = 7.57PSNVNYY194 pKa = 10.33QYY196 pKa = 11.48GFEE199 pKa = 4.28SGVNYY204 pKa = 10.62ANKK207 pKa = 10.24NLGTSAEE214 pKa = 4.24VVEE217 pKa = 4.34IASYY221 pKa = 11.2AGTDD225 pKa = 3.3VTGANVGGNYY235 pKa = 10.07VGTFADD241 pKa = 3.87EE242 pKa = 4.14ATGKK246 pKa = 10.26VVGNALINEE255 pKa = 4.55GCDD258 pKa = 3.16ILFVAAGGSGNGVFTAAKK276 pKa = 8.39EE277 pKa = 3.91ASNVKK282 pKa = 10.5VIGCDD287 pKa = 2.83VDD289 pKa = 4.4QYY291 pKa = 12.01DD292 pKa = 4.75DD293 pKa = 4.02GANGDD298 pKa = 4.13SNIVLTSVLKK308 pKa = 11.26VMDD311 pKa = 4.18KK312 pKa = 10.98NVEE315 pKa = 4.0KK316 pKa = 10.71QLNAVIDD323 pKa = 3.88GTFTGKK329 pKa = 10.1NDD331 pKa = 4.1LLGADD336 pKa = 4.16TDD338 pKa = 3.78STGYY342 pKa = 10.3VKK344 pKa = 10.65QEE346 pKa = 3.54GRR348 pKa = 11.84NQLSADD354 pKa = 4.21TITKK358 pKa = 9.99LDD360 pKa = 3.65EE361 pKa = 4.22VYY363 pKa = 10.97EE364 pKa = 4.03LVKK367 pKa = 11.06NGTVVPAANFNGILPEE383 pKa = 4.35DD384 pKa = 3.87LGG386 pKa = 3.98
MM1 pKa = 7.68KK2 pKa = 10.42KK3 pKa = 9.98RR4 pKa = 11.84VLGLLLCSVMAFSMFAAGCGGSNNAPAPEE33 pKa = 3.91QTEE36 pKa = 4.15NGQNTSTSEE45 pKa = 4.03GGLKK49 pKa = 9.76IAIVSSPSGVDD60 pKa = 3.39DD61 pKa = 5.34GSFNQNNYY69 pKa = 10.33EE70 pKa = 4.37GIQSFIANHH79 pKa = 6.99PDD81 pKa = 3.21ASVTPVKK88 pKa = 10.6EE89 pKa = 4.07EE90 pKa = 4.16SGDD93 pKa = 3.34TAAAVQAVADD103 pKa = 3.93IVADD107 pKa = 3.66YY108 pKa = 11.18NVIVCCGFQFAGIGAITQDD127 pKa = 3.49NPNVNFILVDD137 pKa = 4.03SNPTDD142 pKa = 3.46AQGQEE147 pKa = 3.88INTEE151 pKa = 3.98NIYY154 pKa = 11.54AMTFAEE160 pKa = 4.3QEE162 pKa = 4.14SGFYY166 pKa = 10.56AGVAAALEE174 pKa = 4.35TQSNKK179 pKa = 9.18VAVVNGIAYY188 pKa = 7.57PSNVNYY194 pKa = 10.33QYY196 pKa = 11.48GFEE199 pKa = 4.28SGVNYY204 pKa = 10.62ANKK207 pKa = 10.24NLGTSAEE214 pKa = 4.24VVEE217 pKa = 4.34IASYY221 pKa = 11.2AGTDD225 pKa = 3.3VTGANVGGNYY235 pKa = 10.07VGTFADD241 pKa = 3.87EE242 pKa = 4.14ATGKK246 pKa = 10.26VVGNALINEE255 pKa = 4.55GCDD258 pKa = 3.16ILFVAAGGSGNGVFTAAKK276 pKa = 8.39EE277 pKa = 3.91ASNVKK282 pKa = 10.5VIGCDD287 pKa = 2.83VDD289 pKa = 4.4QYY291 pKa = 12.01DD292 pKa = 4.75DD293 pKa = 4.02GANGDD298 pKa = 4.13SNIVLTSVLKK308 pKa = 11.26VMDD311 pKa = 4.18KK312 pKa = 10.98NVEE315 pKa = 4.0KK316 pKa = 10.71QLNAVIDD323 pKa = 3.88GTFTGKK329 pKa = 10.1NDD331 pKa = 4.1LLGADD336 pKa = 4.16TDD338 pKa = 3.78STGYY342 pKa = 10.3VKK344 pKa = 10.65QEE346 pKa = 3.54GRR348 pKa = 11.84NQLSADD354 pKa = 4.21TITKK358 pKa = 9.99LDD360 pKa = 3.65EE361 pKa = 4.22VYY363 pKa = 10.97EE364 pKa = 4.03LVKK367 pKa = 11.06NGTVVPAANFNGILPEE383 pKa = 4.35DD384 pKa = 3.87LGG386 pKa = 3.98
Molecular weight: 39.91 kDa
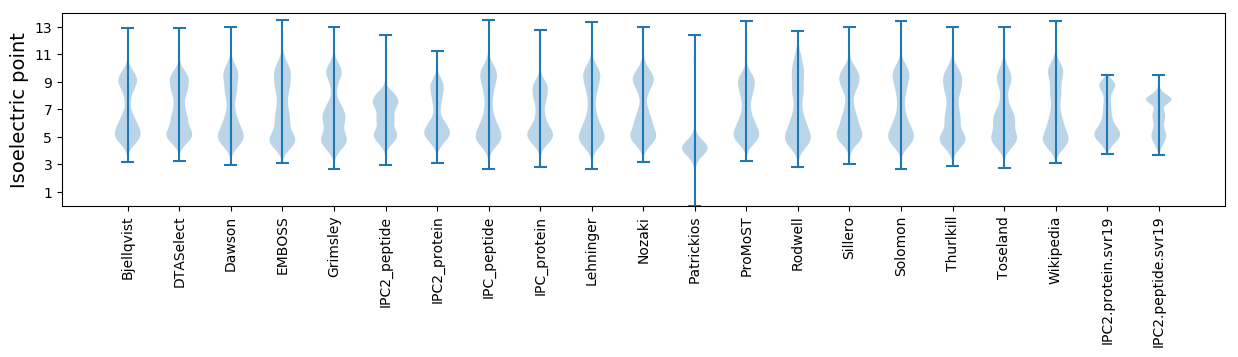
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N1ZMS6|N1ZMS6_9CLOT Uncharacterized protein OS=Clostridium sp. MD294 OX=97138 GN=C820_00687 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835602 |
21 |
2163 |
299.3 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.598 ± 0.053 | 1.423 ± 0.024 |
4.972 ± 0.036 | 7.729 ± 0.054 |
4.224 ± 0.043 | 6.143 ± 0.05 |
1.585 ± 0.019 | 8.9 ± 0.051 |
8.42 ± 0.049 | 8.416 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.839 ± 0.023 | 5.44 ± 0.043 |
2.829 ± 0.026 | 4.372 ± 0.048 |
3.232 ± 0.03 | 5.667 ± 0.037 |
5.858 ± 0.054 | 6.342 ± 0.04 |
0.803 ± 0.015 | 4.208 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
