
Nautilia profundicola (strain ATCC BAA-1463 / DSM 18972 / AmH)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Nautiliales; Nautiliaceae; Nautilia; Nautilia profundicola
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
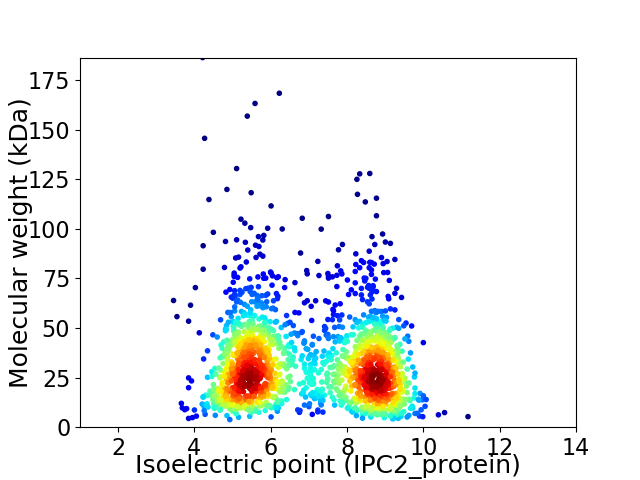
Virtual 2D-PAGE plot for 1730 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B9L754|B9L754_NAUPA NOL1/NOP2/sun family protein OS=Nautilia profundicola (strain ATCC BAA-1463 / DSM 18972 / AmH) OX=598659 GN=NAMH_0021 PE=3 SV=1
MM1 pKa = 6.92KK2 pKa = 10.07TSAKK6 pKa = 10.24VLLSGIAASMILIGCGGGGSSTPSVSGTVEE36 pKa = 3.37ASYY39 pKa = 11.35LKK41 pKa = 10.88GITVCVKK48 pKa = 9.8DD49 pKa = 3.8TSTCATTDD57 pKa = 3.34SNGVFTLNNVSVPVEE72 pKa = 4.13LEE74 pKa = 3.96LKK76 pKa = 10.22IGNSVLADD84 pKa = 3.74LNVSTNNYY92 pKa = 9.4KK93 pKa = 8.67ITPAVLAEE101 pKa = 4.17NNTTIASYY109 pKa = 10.25IGAMLHH115 pKa = 6.98GIAGCDD121 pKa = 3.52LASDD125 pKa = 3.93VCDD128 pKa = 4.75FSSITTVDD136 pKa = 3.1INTSTNLPIITEE148 pKa = 4.34LKK150 pKa = 9.92TILEE154 pKa = 4.5QNSSASISVTVNDD167 pKa = 3.1STMQISEE174 pKa = 4.39VNATLYY180 pKa = 9.7ATEE183 pKa = 4.38NPSMTGTTAYY193 pKa = 10.23SFNGAASAGDD203 pKa = 3.74YY204 pKa = 11.4ASFTYY209 pKa = 10.42NSEE212 pKa = 4.17NNTISYY218 pKa = 9.7QISGNVYY225 pKa = 9.23GNVSGTRR232 pKa = 11.84EE233 pKa = 3.76IEE235 pKa = 3.87NLYY238 pKa = 11.18GNVFFKK244 pKa = 10.88DD245 pKa = 3.65KK246 pKa = 11.19NDD248 pKa = 3.63DD249 pKa = 4.15DD250 pKa = 3.61IFYY253 pKa = 10.32FFSGSLGVAVIPVSDD268 pKa = 3.82TNTSFVVGLQQPDD281 pKa = 3.53KK282 pKa = 11.17NITAEE287 pKa = 4.03DD288 pKa = 3.78LNLIVNKK295 pKa = 9.6KK296 pKa = 8.92YY297 pKa = 11.15NYY299 pKa = 9.76IEE301 pKa = 4.55FDD303 pKa = 3.56TDD305 pKa = 2.87EE306 pKa = 5.03SIYY309 pKa = 10.78FSIIEE314 pKa = 4.11INSTNTADD322 pKa = 6.06LNGTWADD329 pKa = 3.96YY330 pKa = 11.3VDD332 pKa = 3.62QSYY335 pKa = 7.96GTWEE339 pKa = 4.2VNGSHH344 pKa = 7.17LDD346 pKa = 3.48VFDD349 pKa = 4.01ARR351 pKa = 11.84GGKK354 pKa = 8.41IANVIIRR361 pKa = 11.84AGTSRR366 pKa = 11.84AGIVVDD372 pKa = 4.05NVDD375 pKa = 3.11GGFGVGVEE383 pKa = 4.49AKK385 pKa = 10.5ALTDD389 pKa = 3.62ADD391 pKa = 4.17LKK393 pKa = 9.37GTYY396 pKa = 9.51YY397 pKa = 11.23YY398 pKa = 10.87NDD400 pKa = 3.42SGSDD404 pKa = 3.64YY405 pKa = 10.4EE406 pKa = 4.81CYY408 pKa = 9.97GTVTVEE414 pKa = 3.86GTTFKK419 pKa = 11.36YY420 pKa = 10.2KK421 pKa = 10.75DD422 pKa = 3.65EE423 pKa = 4.25WCSDD427 pKa = 3.51NDD429 pKa = 4.05PEE431 pKa = 4.89SGSGTLVLNPTIDD444 pKa = 3.79PDD446 pKa = 4.09QNASTDD452 pKa = 3.57NNITLNGLAQVKK464 pKa = 10.28DD465 pKa = 3.81EE466 pKa = 4.73NGNLTDD472 pKa = 4.0EE473 pKa = 4.95FVFLDD478 pKa = 4.38PDD480 pKa = 3.27SGYY483 pKa = 10.68YY484 pKa = 9.8ISVNIDD490 pKa = 3.31DD491 pKa = 5.12GEE493 pKa = 4.11LSIGSNKK500 pKa = 8.88PLKK503 pKa = 10.53
MM1 pKa = 6.92KK2 pKa = 10.07TSAKK6 pKa = 10.24VLLSGIAASMILIGCGGGGSSTPSVSGTVEE36 pKa = 3.37ASYY39 pKa = 11.35LKK41 pKa = 10.88GITVCVKK48 pKa = 9.8DD49 pKa = 3.8TSTCATTDD57 pKa = 3.34SNGVFTLNNVSVPVEE72 pKa = 4.13LEE74 pKa = 3.96LKK76 pKa = 10.22IGNSVLADD84 pKa = 3.74LNVSTNNYY92 pKa = 9.4KK93 pKa = 8.67ITPAVLAEE101 pKa = 4.17NNTTIASYY109 pKa = 10.25IGAMLHH115 pKa = 6.98GIAGCDD121 pKa = 3.52LASDD125 pKa = 3.93VCDD128 pKa = 4.75FSSITTVDD136 pKa = 3.1INTSTNLPIITEE148 pKa = 4.34LKK150 pKa = 9.92TILEE154 pKa = 4.5QNSSASISVTVNDD167 pKa = 3.1STMQISEE174 pKa = 4.39VNATLYY180 pKa = 9.7ATEE183 pKa = 4.38NPSMTGTTAYY193 pKa = 10.23SFNGAASAGDD203 pKa = 3.74YY204 pKa = 11.4ASFTYY209 pKa = 10.42NSEE212 pKa = 4.17NNTISYY218 pKa = 9.7QISGNVYY225 pKa = 9.23GNVSGTRR232 pKa = 11.84EE233 pKa = 3.76IEE235 pKa = 3.87NLYY238 pKa = 11.18GNVFFKK244 pKa = 10.88DD245 pKa = 3.65KK246 pKa = 11.19NDD248 pKa = 3.63DD249 pKa = 4.15DD250 pKa = 3.61IFYY253 pKa = 10.32FFSGSLGVAVIPVSDD268 pKa = 3.82TNTSFVVGLQQPDD281 pKa = 3.53KK282 pKa = 11.17NITAEE287 pKa = 4.03DD288 pKa = 3.78LNLIVNKK295 pKa = 9.6KK296 pKa = 8.92YY297 pKa = 11.15NYY299 pKa = 9.76IEE301 pKa = 4.55FDD303 pKa = 3.56TDD305 pKa = 2.87EE306 pKa = 5.03SIYY309 pKa = 10.78FSIIEE314 pKa = 4.11INSTNTADD322 pKa = 6.06LNGTWADD329 pKa = 3.96YY330 pKa = 11.3VDD332 pKa = 3.62QSYY335 pKa = 7.96GTWEE339 pKa = 4.2VNGSHH344 pKa = 7.17LDD346 pKa = 3.48VFDD349 pKa = 4.01ARR351 pKa = 11.84GGKK354 pKa = 8.41IANVIIRR361 pKa = 11.84AGTSRR366 pKa = 11.84AGIVVDD372 pKa = 4.05NVDD375 pKa = 3.11GGFGVGVEE383 pKa = 4.49AKK385 pKa = 10.5ALTDD389 pKa = 3.62ADD391 pKa = 4.17LKK393 pKa = 9.37GTYY396 pKa = 9.51YY397 pKa = 11.23YY398 pKa = 10.87NDD400 pKa = 3.42SGSDD404 pKa = 3.64YY405 pKa = 10.4EE406 pKa = 4.81CYY408 pKa = 9.97GTVTVEE414 pKa = 3.86GTTFKK419 pKa = 11.36YY420 pKa = 10.2KK421 pKa = 10.75DD422 pKa = 3.65EE423 pKa = 4.25WCSDD427 pKa = 3.51NDD429 pKa = 4.05PEE431 pKa = 4.89SGSGTLVLNPTIDD444 pKa = 3.79PDD446 pKa = 4.09QNASTDD452 pKa = 3.57NNITLNGLAQVKK464 pKa = 10.28DD465 pKa = 3.81EE466 pKa = 4.73NGNLTDD472 pKa = 4.0EE473 pKa = 4.95FVFLDD478 pKa = 4.38PDD480 pKa = 3.27SGYY483 pKa = 10.68YY484 pKa = 9.8ISVNIDD490 pKa = 3.31DD491 pKa = 5.12GEE493 pKa = 4.11LSIGSNKK500 pKa = 8.88PLKK503 pKa = 10.53
Molecular weight: 53.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B9L9I0|B9L9I0_NAUPA Histidine kinase OS=Nautilia profundicola (strain ATCC BAA-1463 / DSM 18972 / AmH) OX=598659 GN=NAMH_0885 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.35IRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.09RR14 pKa = 11.84THH16 pKa = 6.14GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 8.44TKK25 pKa = 10.35NGRR28 pKa = 11.84KK29 pKa = 8.73VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.35IRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.09RR14 pKa = 11.84THH16 pKa = 6.14GFRR19 pKa = 11.84ARR21 pKa = 11.84LKK23 pKa = 8.44TKK25 pKa = 10.35NGRR28 pKa = 11.84KK29 pKa = 8.73VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
519740 |
37 |
1668 |
300.4 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.241 ± 0.07 | 0.809 ± 0.02 |
5.422 ± 0.047 | 7.658 ± 0.072 |
5.424 ± 0.059 | 5.696 ± 0.061 |
1.566 ± 0.019 | 9.541 ± 0.077 |
10.356 ± 0.086 | 9.458 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.025 | 5.798 ± 0.07 |
3.243 ± 0.032 | 2.312 ± 0.033 |
3.151 ± 0.038 | 5.181 ± 0.042 |
4.57 ± 0.044 | 6.269 ± 0.053 |
0.716 ± 0.018 | 4.262 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
