
Salinimicrobium catena
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Salinimicrobium
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
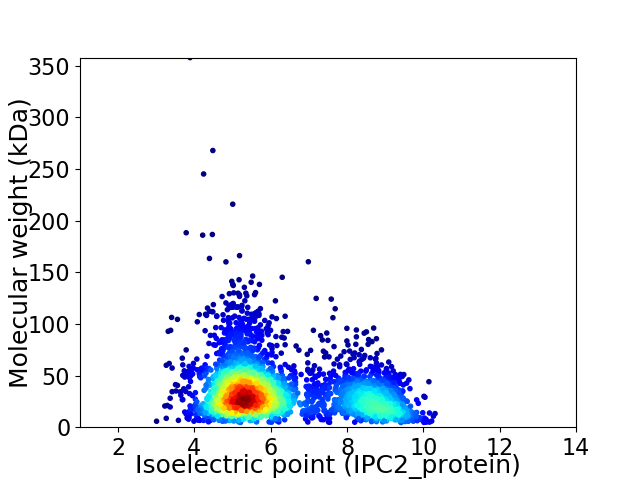
Virtual 2D-PAGE plot for 2965 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
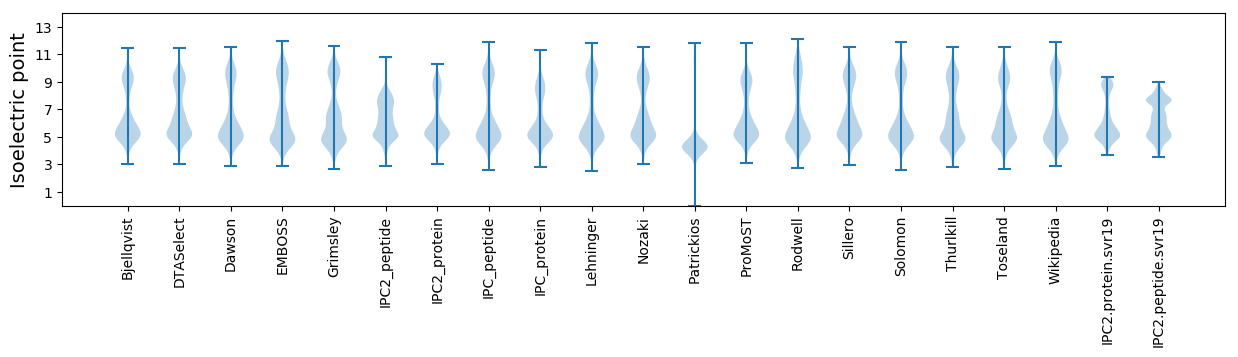
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H5JXK7|A0A1H5JXK7_9FLAO Hemolysin contains CBS domains OS=Salinimicrobium catena OX=390640 GN=SAMN04488034_101899 PE=3 SV=1
MM1 pKa = 7.43EE2 pKa = 5.85NSTKK6 pKa = 10.6HH7 pKa = 5.99LGGAGGKK14 pKa = 8.98LVHH17 pKa = 5.91SVKK20 pKa = 10.83YY21 pKa = 10.5ILIFLLLWVVGCSDD35 pKa = 5.28DD36 pKa = 5.11DD37 pKa = 5.3DD38 pKa = 4.72NVPPPVEE45 pKa = 4.02PTQSIDD51 pKa = 3.45LVLVAEE57 pKa = 4.69NMVSPIGLMPSPDD70 pKa = 3.72DD71 pKa = 3.44TGNLYY76 pKa = 11.04VIDD79 pKa = 4.18QIGKK83 pKa = 8.46VWVIDD88 pKa = 3.83DD89 pKa = 4.08SGNLLEE95 pKa = 5.69EE96 pKa = 4.69PFLDD100 pKa = 3.78VGPMMVTLDD109 pKa = 3.45PGYY112 pKa = 10.42DD113 pKa = 3.35EE114 pKa = 6.13RR115 pKa = 11.84GLLGLAFHH123 pKa = 7.66PDD125 pKa = 3.2YY126 pKa = 11.16SSNGRR131 pKa = 11.84LFVYY135 pKa = 8.11YY136 pKa = 8.3TAPPIAGGPEE146 pKa = 3.99EE147 pKa = 5.5GEE149 pKa = 3.92MWDD152 pKa = 3.28NTSRR156 pKa = 11.84ISEE159 pKa = 4.23FTVSGSNPNQVQAGSEE175 pKa = 4.14KK176 pKa = 10.32IILDD180 pKa = 3.26INQPQFNHH188 pKa = 6.37EE189 pKa = 4.19GGTIAFGPDD198 pKa = 2.8GYY200 pKa = 11.12LYY202 pKa = 10.35ISIGDD207 pKa = 3.75GGASNDD213 pKa = 3.48VAPGHH218 pKa = 6.01VEE220 pKa = 3.44DD221 pKa = 4.97WYY223 pKa = 10.86DD224 pKa = 3.66VNAGGNGQDD233 pKa = 3.69VEE235 pKa = 5.01SNLLGNILRR244 pKa = 11.84IDD246 pKa = 3.53VDD248 pKa = 3.51GGDD251 pKa = 4.04PYY253 pKa = 11.14GIPGDD258 pKa = 3.91NPFVDD263 pKa = 3.09SDD265 pKa = 3.75GRR267 pKa = 11.84DD268 pKa = 3.14EE269 pKa = 4.09IYY271 pKa = 10.46AYY273 pKa = 10.4GFRR276 pKa = 11.84NPFRR280 pKa = 11.84FSFDD284 pKa = 2.67MGGDD288 pKa = 3.15HH289 pKa = 7.52RR290 pKa = 11.84LFVGDD295 pKa = 4.24AGQNLWEE302 pKa = 4.86EE303 pKa = 4.06ISVVEE308 pKa = 4.61NGGNYY313 pKa = 8.44GWNVKK318 pKa = 9.97EE319 pKa = 4.2GTHH322 pKa = 6.87CFDD325 pKa = 4.13ASNNEE330 pKa = 4.05NVLEE334 pKa = 4.24SCPDD338 pKa = 3.12VDD340 pKa = 4.16VLGNEE345 pKa = 4.8LIDD348 pKa = 4.09PVIEE352 pKa = 3.92MSNANNPAGGSTVTIIGGYY371 pKa = 7.34VYY373 pKa = 10.37RR374 pKa = 11.84GTAIPEE380 pKa = 3.82LSGQYY385 pKa = 10.84LFGSFSQDD393 pKa = 3.14FEE395 pKa = 4.4PTGEE399 pKa = 4.13VFFANPSGAGLWDD412 pKa = 3.87FEE414 pKa = 4.19EE415 pKa = 5.94LEE417 pKa = 4.5FKK419 pKa = 9.84STSGDD424 pKa = 2.44IGYY427 pKa = 9.38YY428 pKa = 10.43LKK430 pKa = 11.21GFGQDD435 pKa = 3.37LDD437 pKa = 4.11GEE439 pKa = 4.52IYY441 pKa = 10.39IAVSSMLGPDD451 pKa = 3.52GNTGKK456 pKa = 10.0IFKK459 pKa = 10.24ISLVDD464 pKa = 3.67DD465 pKa = 4.19
MM1 pKa = 7.43EE2 pKa = 5.85NSTKK6 pKa = 10.6HH7 pKa = 5.99LGGAGGKK14 pKa = 8.98LVHH17 pKa = 5.91SVKK20 pKa = 10.83YY21 pKa = 10.5ILIFLLLWVVGCSDD35 pKa = 5.28DD36 pKa = 5.11DD37 pKa = 5.3DD38 pKa = 4.72NVPPPVEE45 pKa = 4.02PTQSIDD51 pKa = 3.45LVLVAEE57 pKa = 4.69NMVSPIGLMPSPDD70 pKa = 3.72DD71 pKa = 3.44TGNLYY76 pKa = 11.04VIDD79 pKa = 4.18QIGKK83 pKa = 8.46VWVIDD88 pKa = 3.83DD89 pKa = 4.08SGNLLEE95 pKa = 5.69EE96 pKa = 4.69PFLDD100 pKa = 3.78VGPMMVTLDD109 pKa = 3.45PGYY112 pKa = 10.42DD113 pKa = 3.35EE114 pKa = 6.13RR115 pKa = 11.84GLLGLAFHH123 pKa = 7.66PDD125 pKa = 3.2YY126 pKa = 11.16SSNGRR131 pKa = 11.84LFVYY135 pKa = 8.11YY136 pKa = 8.3TAPPIAGGPEE146 pKa = 3.99EE147 pKa = 5.5GEE149 pKa = 3.92MWDD152 pKa = 3.28NTSRR156 pKa = 11.84ISEE159 pKa = 4.23FTVSGSNPNQVQAGSEE175 pKa = 4.14KK176 pKa = 10.32IILDD180 pKa = 3.26INQPQFNHH188 pKa = 6.37EE189 pKa = 4.19GGTIAFGPDD198 pKa = 2.8GYY200 pKa = 11.12LYY202 pKa = 10.35ISIGDD207 pKa = 3.75GGASNDD213 pKa = 3.48VAPGHH218 pKa = 6.01VEE220 pKa = 3.44DD221 pKa = 4.97WYY223 pKa = 10.86DD224 pKa = 3.66VNAGGNGQDD233 pKa = 3.69VEE235 pKa = 5.01SNLLGNILRR244 pKa = 11.84IDD246 pKa = 3.53VDD248 pKa = 3.51GGDD251 pKa = 4.04PYY253 pKa = 11.14GIPGDD258 pKa = 3.91NPFVDD263 pKa = 3.09SDD265 pKa = 3.75GRR267 pKa = 11.84DD268 pKa = 3.14EE269 pKa = 4.09IYY271 pKa = 10.46AYY273 pKa = 10.4GFRR276 pKa = 11.84NPFRR280 pKa = 11.84FSFDD284 pKa = 2.67MGGDD288 pKa = 3.15HH289 pKa = 7.52RR290 pKa = 11.84LFVGDD295 pKa = 4.24AGQNLWEE302 pKa = 4.86EE303 pKa = 4.06ISVVEE308 pKa = 4.61NGGNYY313 pKa = 8.44GWNVKK318 pKa = 9.97EE319 pKa = 4.2GTHH322 pKa = 6.87CFDD325 pKa = 4.13ASNNEE330 pKa = 4.05NVLEE334 pKa = 4.24SCPDD338 pKa = 3.12VDD340 pKa = 4.16VLGNEE345 pKa = 4.8LIDD348 pKa = 4.09PVIEE352 pKa = 3.92MSNANNPAGGSTVTIIGGYY371 pKa = 7.34VYY373 pKa = 10.37RR374 pKa = 11.84GTAIPEE380 pKa = 3.82LSGQYY385 pKa = 10.84LFGSFSQDD393 pKa = 3.14FEE395 pKa = 4.4PTGEE399 pKa = 4.13VFFANPSGAGLWDD412 pKa = 3.87FEE414 pKa = 4.19EE415 pKa = 5.94LEE417 pKa = 4.5FKK419 pKa = 9.84STSGDD424 pKa = 2.44IGYY427 pKa = 9.38YY428 pKa = 10.43LKK430 pKa = 11.21GFGQDD435 pKa = 3.37LDD437 pKa = 4.11GEE439 pKa = 4.52IYY441 pKa = 10.39IAVSSMLGPDD451 pKa = 3.52GNTGKK456 pKa = 10.0IFKK459 pKa = 10.24ISLVDD464 pKa = 3.67DD465 pKa = 4.19
Molecular weight: 50.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H5JB70|A0A1H5JB70_9FLAO Uncharacterized protein OS=Salinimicrobium catena OX=390640 GN=SAMN04488034_101685 PE=4 SV=1
MM1 pKa = 7.22FSIGRR6 pKa = 11.84LFCCYY11 pKa = 9.94FGHH14 pKa = 7.23RR15 pKa = 11.84LHH17 pKa = 7.04ISRR20 pKa = 11.84RR21 pKa = 11.84ITNHH25 pKa = 4.96IKK27 pKa = 10.11EE28 pKa = 4.37YY29 pKa = 10.18SCTRR33 pKa = 11.84CGQEE37 pKa = 3.65MTDD40 pKa = 3.4TANGFLANLTPRR52 pKa = 11.84LQEE55 pKa = 4.08TNTFLAKK62 pKa = 9.19IHH64 pKa = 5.2EE65 pKa = 4.3RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84NRR70 pKa = 11.84KK71 pKa = 8.52MILKK75 pKa = 10.42ASS77 pKa = 3.67
MM1 pKa = 7.22FSIGRR6 pKa = 11.84LFCCYY11 pKa = 9.94FGHH14 pKa = 7.23RR15 pKa = 11.84LHH17 pKa = 7.04ISRR20 pKa = 11.84RR21 pKa = 11.84ITNHH25 pKa = 4.96IKK27 pKa = 10.11EE28 pKa = 4.37YY29 pKa = 10.18SCTRR33 pKa = 11.84CGQEE37 pKa = 3.65MTDD40 pKa = 3.4TANGFLANLTPRR52 pKa = 11.84LQEE55 pKa = 4.08TNTFLAKK62 pKa = 9.19IHH64 pKa = 5.2EE65 pKa = 4.3RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84NRR70 pKa = 11.84KK71 pKa = 8.52MILKK75 pKa = 10.42ASS77 pKa = 3.67
Molecular weight: 9.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981291 |
39 |
3347 |
331.0 |
37.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.722 ± 0.045 | 0.653 ± 0.015 |
5.439 ± 0.034 | 7.928 ± 0.053 |
5.189 ± 0.036 | 6.653 ± 0.042 |
1.856 ± 0.021 | 7.039 ± 0.037 |
7.29 ± 0.054 | 9.607 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.022 | 5.168 ± 0.033 |
3.703 ± 0.022 | 3.477 ± 0.022 |
4.152 ± 0.026 | 6.156 ± 0.031 |
5.236 ± 0.039 | 6.442 ± 0.033 |
1.089 ± 0.018 | 3.877 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
