
Salinivibrio sp. KP-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Salinivibrio; unclassified Salinivibrio
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
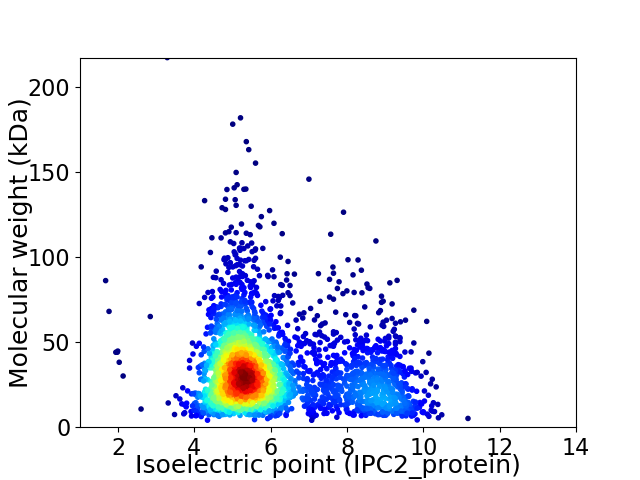
Virtual 2D-PAGE plot for 3027 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F5AU70|A0A0F5AU70_9GAMM Peptidase M13 OS=Salinivibrio sp. KP-1 OX=1406902 GN=WN56_00080 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.43KK3 pKa = 9.02TILAMAVPALLAAGSASASINLYY26 pKa = 9.87DD27 pKa = 3.95AEE29 pKa = 4.44GVKK32 pKa = 10.3VDD34 pKa = 3.57ASGAAEE40 pKa = 3.93VQYY43 pKa = 10.43FQAIGEE49 pKa = 4.37NKK51 pKa = 10.09DD52 pKa = 3.46GEE54 pKa = 4.32WRR56 pKa = 11.84LDD58 pKa = 3.83DD59 pKa = 5.06GDD61 pKa = 3.97FAVNTEE67 pKa = 4.22VAVSEE72 pKa = 4.05DD73 pKa = 3.32LAAIGHH79 pKa = 6.11FAFKK83 pKa = 10.9FEE85 pKa = 5.95DD86 pKa = 3.68STTKK90 pKa = 10.58NDD92 pKa = 3.17EE93 pKa = 4.34LYY95 pKa = 11.2VGFKK99 pKa = 10.7GSDD102 pKa = 2.59WGTLTFGRR110 pKa = 11.84QLLINDD116 pKa = 4.16DD117 pKa = 3.54MGITKK122 pKa = 10.19DD123 pKa = 3.89YY124 pKa = 10.2EE125 pKa = 4.0LSSASNGTLRR135 pKa = 11.84TEE137 pKa = 4.72GDD139 pKa = 3.33QVAKK143 pKa = 10.46YY144 pKa = 10.51VYY146 pKa = 10.88DD147 pKa = 3.42NGTFYY152 pKa = 11.22AGLSTLQNSDD162 pKa = 3.52GEE164 pKa = 4.4EE165 pKa = 3.94SSDD168 pKa = 3.49EE169 pKa = 3.88YY170 pKa = 11.12KK171 pKa = 10.46IYY173 pKa = 10.49DD174 pKa = 3.17GRR176 pKa = 11.84LGYY179 pKa = 10.17RR180 pKa = 11.84VADD183 pKa = 3.01VDD185 pKa = 3.57MRR187 pKa = 11.84VYY189 pKa = 10.73YY190 pKa = 10.43YY191 pKa = 10.68YY192 pKa = 11.34ADD194 pKa = 3.96DD195 pKa = 4.97AGVNTAGDD203 pKa = 3.87DD204 pKa = 3.48FVYY207 pKa = 10.65ASKK210 pKa = 11.45GDD212 pKa = 3.53VEE214 pKa = 4.54NYY216 pKa = 10.06NFEE219 pKa = 4.04VEE221 pKa = 4.24YY222 pKa = 11.09AGIEE226 pKa = 4.23NVSLAAAYY234 pKa = 9.08GVNTVDD240 pKa = 3.61TGSSDD245 pKa = 3.37TEE247 pKa = 3.82ADD249 pKa = 3.96FFSVAADD256 pKa = 3.62YY257 pKa = 9.45THH259 pKa = 6.99GKK261 pKa = 4.82TTYY264 pKa = 10.86AIGIDD269 pKa = 3.6NKK271 pKa = 11.37DD272 pKa = 3.31NDD274 pKa = 3.8NEE276 pKa = 4.09EE277 pKa = 4.84DD278 pKa = 3.6VTGYY282 pKa = 9.43YY283 pKa = 11.12ANVTYY288 pKa = 10.86ALHH291 pKa = 6.63NNAKK295 pKa = 9.88VYY297 pKa = 11.25AEE299 pKa = 4.34VGDD302 pKa = 4.05NDD304 pKa = 4.92ADD306 pKa = 4.06NNDD309 pKa = 3.23WGYY312 pKa = 9.36VAGMEE317 pKa = 4.59VKK319 pKa = 10.61FF320 pKa = 4.19
MM1 pKa = 7.68KK2 pKa = 9.43KK3 pKa = 9.02TILAMAVPALLAAGSASASINLYY26 pKa = 9.87DD27 pKa = 3.95AEE29 pKa = 4.44GVKK32 pKa = 10.3VDD34 pKa = 3.57ASGAAEE40 pKa = 3.93VQYY43 pKa = 10.43FQAIGEE49 pKa = 4.37NKK51 pKa = 10.09DD52 pKa = 3.46GEE54 pKa = 4.32WRR56 pKa = 11.84LDD58 pKa = 3.83DD59 pKa = 5.06GDD61 pKa = 3.97FAVNTEE67 pKa = 4.22VAVSEE72 pKa = 4.05DD73 pKa = 3.32LAAIGHH79 pKa = 6.11FAFKK83 pKa = 10.9FEE85 pKa = 5.95DD86 pKa = 3.68STTKK90 pKa = 10.58NDD92 pKa = 3.17EE93 pKa = 4.34LYY95 pKa = 11.2VGFKK99 pKa = 10.7GSDD102 pKa = 2.59WGTLTFGRR110 pKa = 11.84QLLINDD116 pKa = 4.16DD117 pKa = 3.54MGITKK122 pKa = 10.19DD123 pKa = 3.89YY124 pKa = 10.2EE125 pKa = 4.0LSSASNGTLRR135 pKa = 11.84TEE137 pKa = 4.72GDD139 pKa = 3.33QVAKK143 pKa = 10.46YY144 pKa = 10.51VYY146 pKa = 10.88DD147 pKa = 3.42NGTFYY152 pKa = 11.22AGLSTLQNSDD162 pKa = 3.52GEE164 pKa = 4.4EE165 pKa = 3.94SSDD168 pKa = 3.49EE169 pKa = 3.88YY170 pKa = 11.12KK171 pKa = 10.46IYY173 pKa = 10.49DD174 pKa = 3.17GRR176 pKa = 11.84LGYY179 pKa = 10.17RR180 pKa = 11.84VADD183 pKa = 3.01VDD185 pKa = 3.57MRR187 pKa = 11.84VYY189 pKa = 10.73YY190 pKa = 10.43YY191 pKa = 10.68YY192 pKa = 11.34ADD194 pKa = 3.96DD195 pKa = 4.97AGVNTAGDD203 pKa = 3.87DD204 pKa = 3.48FVYY207 pKa = 10.65ASKK210 pKa = 11.45GDD212 pKa = 3.53VEE214 pKa = 4.54NYY216 pKa = 10.06NFEE219 pKa = 4.04VEE221 pKa = 4.24YY222 pKa = 11.09AGIEE226 pKa = 4.23NVSLAAAYY234 pKa = 9.08GVNTVDD240 pKa = 3.61TGSSDD245 pKa = 3.37TEE247 pKa = 3.82ADD249 pKa = 3.96FFSVAADD256 pKa = 3.62YY257 pKa = 9.45THH259 pKa = 6.99GKK261 pKa = 4.82TTYY264 pKa = 10.86AIGIDD269 pKa = 3.6NKK271 pKa = 11.37DD272 pKa = 3.31NDD274 pKa = 3.8NEE276 pKa = 4.09EE277 pKa = 4.84DD278 pKa = 3.6VTGYY282 pKa = 9.43YY283 pKa = 11.12ANVTYY288 pKa = 10.86ALHH291 pKa = 6.63NNAKK295 pKa = 9.88VYY297 pKa = 11.25AEE299 pKa = 4.34VGDD302 pKa = 4.05NDD304 pKa = 4.92ADD306 pKa = 4.06NNDD309 pKa = 3.23WGYY312 pKa = 9.36VAGMEE317 pKa = 4.59VKK319 pKa = 10.61FF320 pKa = 4.19
Molecular weight: 34.79 kDa
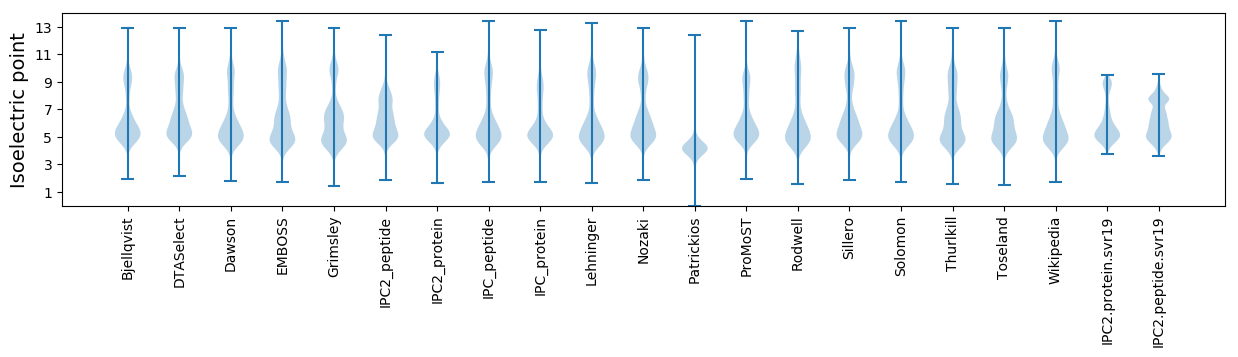
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F5AQ48|A0A0F5AQ48_9GAMM DNA polymerase III subunit psi OS=Salinivibrio sp. KP-1 OX=1406902 GN=WN56_11480 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.44VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 8.23QLSKK44 pKa = 11.23
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.44VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 8.23QLSKK44 pKa = 11.23
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
976084 |
35 |
2095 |
322.5 |
35.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.92 ± 0.05 | 1.014 ± 0.015 |
5.943 ± 0.045 | 5.812 ± 0.037 |
3.735 ± 0.028 | 7.022 ± 0.041 |
2.521 ± 0.026 | 5.555 ± 0.033 |
4.297 ± 0.033 | 10.362 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.023 | 3.525 ± 0.025 |
4.257 ± 0.025 | 5.051 ± 0.041 |
5.308 ± 0.034 | 6.013 ± 0.036 |
5.508 ± 0.034 | 7.249 ± 0.038 |
1.324 ± 0.018 | 2.879 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
