
Lachnospiraceae bacterium NK3A20
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
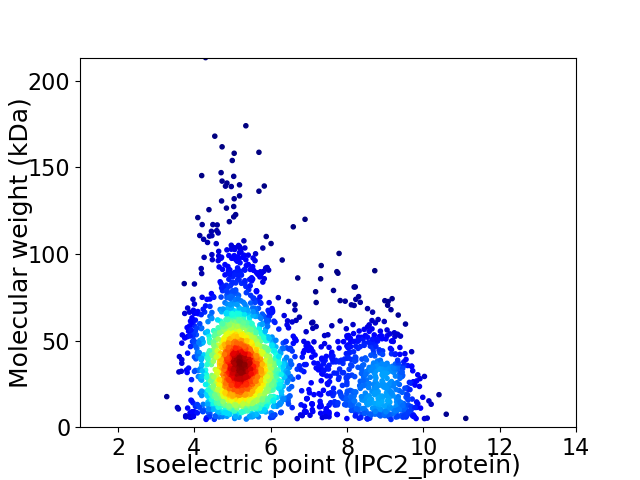
Virtual 2D-PAGE plot for 2503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3ZQ01|A0A1H3ZQ01_9FIRM CxxC motif-containing protein OS=Lachnospiraceae bacterium NK3A20 OX=877406 GN=SAMN02745687_02306 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.36KK3 pKa = 9.79RR4 pKa = 11.84VLSIVLGTAMAASVLAGCGSTDD26 pKa = 3.25SAEE29 pKa = 4.41PAATAAPADD38 pKa = 4.13ADD40 pKa = 4.02TAATEE45 pKa = 4.14AATDD49 pKa = 3.62AATTAAEE56 pKa = 4.67GNTEE60 pKa = 4.07TATTEE65 pKa = 3.72AAADD69 pKa = 3.93AEE71 pKa = 4.63AGDD74 pKa = 3.95IPGGRR79 pKa = 11.84VYY81 pKa = 11.22LLNFKK86 pKa = 10.66PEE88 pKa = 4.19TDD90 pKa = 5.07DD91 pKa = 3.36AWQDD95 pKa = 3.39LASTYY100 pKa = 11.25NGMGGNVTVVTAADD114 pKa = 3.7GQYY117 pKa = 9.85ATTLQSEE124 pKa = 4.66MAKK127 pKa = 10.15SEE129 pKa = 3.9APTIFNIGNSTDD141 pKa = 3.07AQTWDD146 pKa = 4.09DD147 pKa = 3.6YY148 pKa = 11.75TYY150 pKa = 10.95DD151 pKa = 3.59LKK153 pKa = 11.49GSAVYY158 pKa = 10.55DD159 pKa = 3.68HH160 pKa = 7.07VSDD163 pKa = 4.17HH164 pKa = 6.66SLDD167 pKa = 3.37VLYY170 pKa = 10.82NGKK173 pKa = 7.7TAAIANCYY181 pKa = 7.38EE182 pKa = 4.23AYY184 pKa = 10.45GIIYY188 pKa = 10.35NKK190 pKa = 9.8TILDD194 pKa = 4.44AYY196 pKa = 8.99CTLDD200 pKa = 3.36GAKK203 pKa = 9.85VASADD208 pKa = 3.97EE209 pKa = 4.27IQDD212 pKa = 3.42FDD214 pKa = 4.1TLKK217 pKa = 11.06AVADD221 pKa = 4.95DD222 pKa = 3.44INSRR226 pKa = 11.84VDD228 pKa = 3.63EE229 pKa = 4.64LNEE232 pKa = 4.07ALAGTAFDD240 pKa = 4.01GTVTEE245 pKa = 4.92AFASAGLDD253 pKa = 3.7DD254 pKa = 4.15SSSWRR259 pKa = 11.84FSGHH263 pKa = 6.45LANFPLYY270 pKa = 11.0YY271 pKa = 10.18EE272 pKa = 4.65FKK274 pKa = 10.8DD275 pKa = 5.16DD276 pKa = 4.37GLEE279 pKa = 4.15DD280 pKa = 3.64LTAGEE285 pKa = 4.53PEE287 pKa = 4.03IKK289 pKa = 9.38GTYY292 pKa = 9.03LDD294 pKa = 3.85QYY296 pKa = 9.73QKK298 pKa = 10.65VWDD301 pKa = 4.01MYY303 pKa = 10.82VADD306 pKa = 4.45SAADD310 pKa = 3.65PATLAGGTYY319 pKa = 9.73NAEE322 pKa = 4.12EE323 pKa = 3.96EE324 pKa = 4.53LGLGEE329 pKa = 4.94AVFYY333 pKa = 11.32QNGDD337 pKa = 3.32WEE339 pKa = 5.13FSALTNPEE347 pKa = 3.17NGYY350 pKa = 10.03IVTADD355 pKa = 4.8DD356 pKa = 4.82LDD358 pKa = 3.58MMPIYY363 pKa = 10.58FGVDD367 pKa = 3.36DD368 pKa = 4.76EE369 pKa = 5.0NEE371 pKa = 4.0GLAVGTEE378 pKa = 4.01NHH380 pKa = 5.77WAVNSKK386 pKa = 10.08AAQEE390 pKa = 5.11DD391 pKa = 3.7IDD393 pKa = 4.11ASLYY397 pKa = 9.18FLNWAITSDD406 pKa = 3.64EE407 pKa = 3.99GRR409 pKa = 11.84DD410 pKa = 3.67AIVNTMGLSAPFDD423 pKa = 3.75TFTGDD428 pKa = 3.9FAPANAFGAKK438 pKa = 9.49AAVYY442 pKa = 9.07TEE444 pKa = 4.11AGKK447 pKa = 8.71TSTAWSFNATPNVDD461 pKa = 3.0DD462 pKa = 3.57WRR464 pKa = 11.84ADD466 pKa = 3.45VVSALTAYY474 pKa = 9.73TDD476 pKa = 4.83GSGDD480 pKa = 3.08WDD482 pKa = 3.63AVKK485 pKa = 10.64DD486 pKa = 3.81AFVTGWADD494 pKa = 3.23QWRR497 pKa = 11.84LAHH500 pKa = 7.14AEE502 pKa = 3.9
MM1 pKa = 7.73KK2 pKa = 10.36KK3 pKa = 9.79RR4 pKa = 11.84VLSIVLGTAMAASVLAGCGSTDD26 pKa = 3.25SAEE29 pKa = 4.41PAATAAPADD38 pKa = 4.13ADD40 pKa = 4.02TAATEE45 pKa = 4.14AATDD49 pKa = 3.62AATTAAEE56 pKa = 4.67GNTEE60 pKa = 4.07TATTEE65 pKa = 3.72AAADD69 pKa = 3.93AEE71 pKa = 4.63AGDD74 pKa = 3.95IPGGRR79 pKa = 11.84VYY81 pKa = 11.22LLNFKK86 pKa = 10.66PEE88 pKa = 4.19TDD90 pKa = 5.07DD91 pKa = 3.36AWQDD95 pKa = 3.39LASTYY100 pKa = 11.25NGMGGNVTVVTAADD114 pKa = 3.7GQYY117 pKa = 9.85ATTLQSEE124 pKa = 4.66MAKK127 pKa = 10.15SEE129 pKa = 3.9APTIFNIGNSTDD141 pKa = 3.07AQTWDD146 pKa = 4.09DD147 pKa = 3.6YY148 pKa = 11.75TYY150 pKa = 10.95DD151 pKa = 3.59LKK153 pKa = 11.49GSAVYY158 pKa = 10.55DD159 pKa = 3.68HH160 pKa = 7.07VSDD163 pKa = 4.17HH164 pKa = 6.66SLDD167 pKa = 3.37VLYY170 pKa = 10.82NGKK173 pKa = 7.7TAAIANCYY181 pKa = 7.38EE182 pKa = 4.23AYY184 pKa = 10.45GIIYY188 pKa = 10.35NKK190 pKa = 9.8TILDD194 pKa = 4.44AYY196 pKa = 8.99CTLDD200 pKa = 3.36GAKK203 pKa = 9.85VASADD208 pKa = 3.97EE209 pKa = 4.27IQDD212 pKa = 3.42FDD214 pKa = 4.1TLKK217 pKa = 11.06AVADD221 pKa = 4.95DD222 pKa = 3.44INSRR226 pKa = 11.84VDD228 pKa = 3.63EE229 pKa = 4.64LNEE232 pKa = 4.07ALAGTAFDD240 pKa = 4.01GTVTEE245 pKa = 4.92AFASAGLDD253 pKa = 3.7DD254 pKa = 4.15SSSWRR259 pKa = 11.84FSGHH263 pKa = 6.45LANFPLYY270 pKa = 11.0YY271 pKa = 10.18EE272 pKa = 4.65FKK274 pKa = 10.8DD275 pKa = 5.16DD276 pKa = 4.37GLEE279 pKa = 4.15DD280 pKa = 3.64LTAGEE285 pKa = 4.53PEE287 pKa = 4.03IKK289 pKa = 9.38GTYY292 pKa = 9.03LDD294 pKa = 3.85QYY296 pKa = 9.73QKK298 pKa = 10.65VWDD301 pKa = 4.01MYY303 pKa = 10.82VADD306 pKa = 4.45SAADD310 pKa = 3.65PATLAGGTYY319 pKa = 9.73NAEE322 pKa = 4.12EE323 pKa = 3.96EE324 pKa = 4.53LGLGEE329 pKa = 4.94AVFYY333 pKa = 11.32QNGDD337 pKa = 3.32WEE339 pKa = 5.13FSALTNPEE347 pKa = 3.17NGYY350 pKa = 10.03IVTADD355 pKa = 4.8DD356 pKa = 4.82LDD358 pKa = 3.58MMPIYY363 pKa = 10.58FGVDD367 pKa = 3.36DD368 pKa = 4.76EE369 pKa = 5.0NEE371 pKa = 4.0GLAVGTEE378 pKa = 4.01NHH380 pKa = 5.77WAVNSKK386 pKa = 10.08AAQEE390 pKa = 5.11DD391 pKa = 3.7IDD393 pKa = 4.11ASLYY397 pKa = 9.18FLNWAITSDD406 pKa = 3.64EE407 pKa = 3.99GRR409 pKa = 11.84DD410 pKa = 3.67AIVNTMGLSAPFDD423 pKa = 3.75TFTGDD428 pKa = 3.9FAPANAFGAKK438 pKa = 9.49AAVYY442 pKa = 9.07TEE444 pKa = 4.11AGKK447 pKa = 8.71TSTAWSFNATPNVDD461 pKa = 3.0DD462 pKa = 3.57WRR464 pKa = 11.84ADD466 pKa = 3.45VVSALTAYY474 pKa = 9.73TDD476 pKa = 4.83GSGDD480 pKa = 3.08WDD482 pKa = 3.63AVKK485 pKa = 10.64DD486 pKa = 3.81AFVTGWADD494 pKa = 3.23QWRR497 pKa = 11.84LAHH500 pKa = 7.14AEE502 pKa = 3.9
Molecular weight: 53.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3WT17|A0A1H3WT17_9FIRM Uncharacterized protein PH0010 family/AmmeMemoRadiSam system protein A/AmmeMemoRadiSam system protein B OS=Lachnospiraceae bacterium NK3A20 OX=877406 GN=SAMN02745687_01019 PE=4 SV=1
MM1 pKa = 7.64SKK3 pKa = 7.87MTLQPKK9 pKa = 9.35KK10 pKa = 8.73LQRR13 pKa = 11.84SRR15 pKa = 11.84VHH17 pKa = 6.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.38AGRR29 pKa = 11.84KK30 pKa = 8.69VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.26GRR40 pKa = 11.84AKK42 pKa = 10.1LTVV45 pKa = 3.04
MM1 pKa = 7.64SKK3 pKa = 7.87MTLQPKK9 pKa = 9.35KK10 pKa = 8.73LQRR13 pKa = 11.84SRR15 pKa = 11.84VHH17 pKa = 6.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.38AGRR29 pKa = 11.84KK30 pKa = 8.69VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.26GRR40 pKa = 11.84AKK42 pKa = 10.1LTVV45 pKa = 3.04
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
869147 |
39 |
1981 |
347.2 |
38.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.965 ± 0.059 | 1.298 ± 0.018 |
6.285 ± 0.044 | 6.951 ± 0.053 |
3.958 ± 0.034 | 7.455 ± 0.041 |
1.984 ± 0.021 | 6.718 ± 0.044 |
5.216 ± 0.042 | 8.772 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.893 ± 0.022 | 3.98 ± 0.029 |
3.709 ± 0.026 | 3.273 ± 0.029 |
5.418 ± 0.043 | 5.713 ± 0.037 |
5.736 ± 0.041 | 6.625 ± 0.04 |
1.043 ± 0.018 | 4.006 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
