
Candidatus Synechococcus spongiarum LMB bulk10D
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; Candidatus Synechococcus spongiarum
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
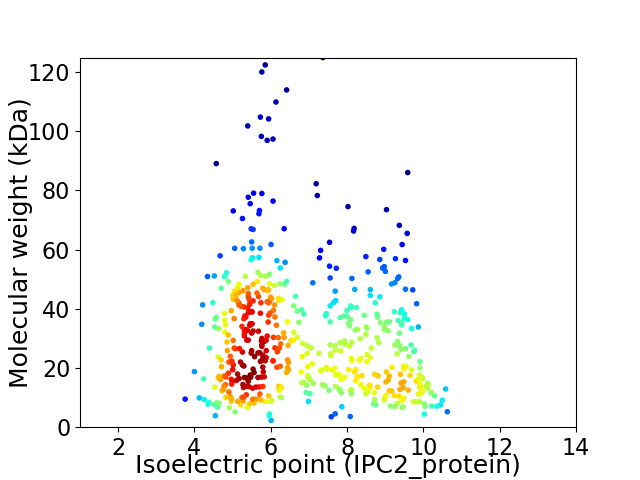
Virtual 2D-PAGE plot for 532 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
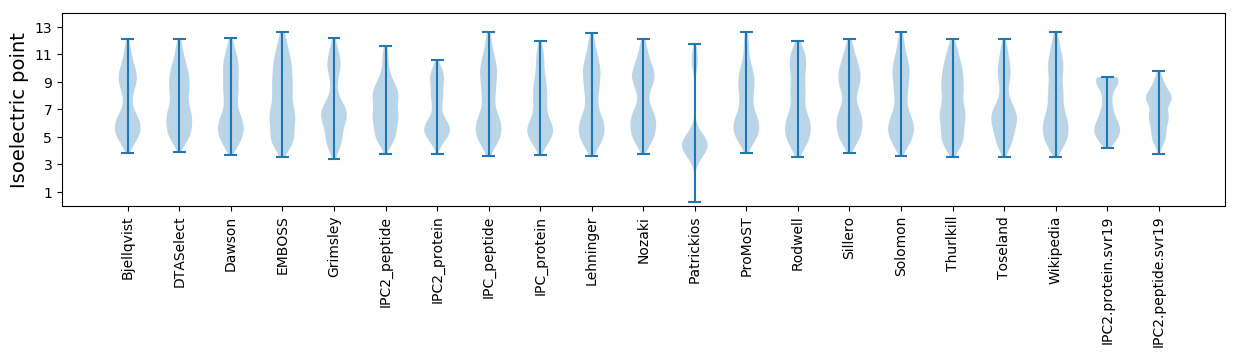
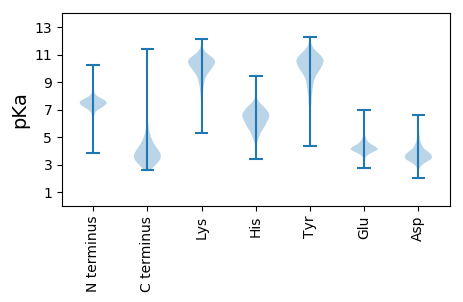
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N4ASP3|A0A6N4ASP3_9SYNE Transcriptional repressor OS=Candidatus Synechococcus spongiarum LMB bulk10D OX=1449121 GN=BO98_00165 PE=3 SV=1
MM1 pKa = 7.57AVGIYY6 pKa = 9.88FATTTGKK13 pKa = 7.71TQDD16 pKa = 3.25VAEE19 pKa = 4.63RR20 pKa = 11.84LHH22 pKa = 6.83ALLSSANAPKK32 pKa = 10.59DD33 pKa = 3.69LADD36 pKa = 3.67MADD39 pKa = 3.09VSEE42 pKa = 4.19FTEE45 pKa = 4.96LDD47 pKa = 3.48GLICGVPTWNTGAEE61 pKa = 4.26TEE63 pKa = 4.27RR64 pKa = 11.84SGTAWDD70 pKa = 4.6GVLEE74 pKa = 5.3DD75 pKa = 4.93IAEE78 pKa = 4.36LDD80 pKa = 3.86LNGKK84 pKa = 8.98FVAVFGLGDD93 pKa = 3.59SGSYY97 pKa = 9.69TDD99 pKa = 4.21NFCDD103 pKa = 5.08AMEE106 pKa = 4.29EE107 pKa = 3.89LHH109 pKa = 7.04NFFKK113 pKa = 10.87QAGATMVGYY122 pKa = 9.12MDD124 pKa = 3.69KK125 pKa = 11.21SNYY128 pKa = 7.86TFEE131 pKa = 4.23EE132 pKa = 4.42SKK134 pKa = 11.06SVIGDD139 pKa = 4.01KK140 pKa = 10.9FCGLALDD147 pKa = 4.73EE148 pKa = 5.47DD149 pKa = 4.75SEE151 pKa = 5.92SDD153 pKa = 3.43MTDD156 pKa = 2.95DD157 pKa = 6.22RR158 pKa = 11.84LAKK161 pKa = 9.48WAEE164 pKa = 3.86QLKK167 pKa = 10.13QEE169 pKa = 4.6IPGLL173 pKa = 3.83
MM1 pKa = 7.57AVGIYY6 pKa = 9.88FATTTGKK13 pKa = 7.71TQDD16 pKa = 3.25VAEE19 pKa = 4.63RR20 pKa = 11.84LHH22 pKa = 6.83ALLSSANAPKK32 pKa = 10.59DD33 pKa = 3.69LADD36 pKa = 3.67MADD39 pKa = 3.09VSEE42 pKa = 4.19FTEE45 pKa = 4.96LDD47 pKa = 3.48GLICGVPTWNTGAEE61 pKa = 4.26TEE63 pKa = 4.27RR64 pKa = 11.84SGTAWDD70 pKa = 4.6GVLEE74 pKa = 5.3DD75 pKa = 4.93IAEE78 pKa = 4.36LDD80 pKa = 3.86LNGKK84 pKa = 8.98FVAVFGLGDD93 pKa = 3.59SGSYY97 pKa = 9.69TDD99 pKa = 4.21NFCDD103 pKa = 5.08AMEE106 pKa = 4.29EE107 pKa = 3.89LHH109 pKa = 7.04NFFKK113 pKa = 10.87QAGATMVGYY122 pKa = 9.12MDD124 pKa = 3.69KK125 pKa = 11.21SNYY128 pKa = 7.86TFEE131 pKa = 4.23EE132 pKa = 4.42SKK134 pKa = 11.06SVIGDD139 pKa = 4.01KK140 pKa = 10.9FCGLALDD147 pKa = 4.73EE148 pKa = 5.47DD149 pKa = 4.75SEE151 pKa = 5.92SDD153 pKa = 3.43MTDD156 pKa = 2.95DD157 pKa = 6.22RR158 pKa = 11.84LAKK161 pKa = 9.48WAEE164 pKa = 3.86QLKK167 pKa = 10.13QEE169 pKa = 4.6IPGLL173 pKa = 3.83
Molecular weight: 18.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N4ATU9|A0A6N4ATU9_9SYNE Rubredoxin OS=Candidatus Synechococcus spongiarum LMB bulk10D OX=1449121 GN=BO98_02760 PE=3 SV=1
MM1 pKa = 7.15ITTASRR7 pKa = 11.84QSALEE12 pKa = 3.79RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.38ALATAGKK22 pKa = 9.61AAASRR27 pKa = 11.84YY28 pKa = 9.97AKK30 pKa = 9.37TRR32 pKa = 11.84QRR34 pKa = 11.84NISDD38 pKa = 3.46EE39 pKa = 3.87QSPRR43 pKa = 11.84RR44 pKa = 11.84GHH46 pKa = 5.22VVTRR50 pKa = 11.84TVSVTPRR57 pKa = 11.84PRR59 pKa = 11.84PTVVFSPAAVAQPAPAGTPNLNTRR83 pKa = 11.84KK84 pKa = 9.65ISNPTRR90 pKa = 11.84DD91 pKa = 3.05LALARR96 pKa = 11.84RR97 pKa = 11.84QALSRR102 pKa = 11.84QGKK105 pKa = 10.08RR106 pKa = 11.84NDD108 pKa = 3.53PGNNRR113 pKa = 11.84IRR115 pKa = 11.84TEE117 pKa = 3.49VRR119 pKa = 11.84RR120 pKa = 11.84PSRR123 pKa = 11.84TTPQPAVPDD132 pKa = 3.88RR133 pKa = 11.84QDD135 pKa = 3.05TSHH138 pKa = 6.61TCDD141 pKa = 3.01CRR143 pKa = 11.84KK144 pKa = 10.16RR145 pKa = 11.84PGKK148 pKa = 8.41EE149 pKa = 3.43VRR151 pKa = 11.84KK152 pKa = 9.71PNLTSLSTGRR162 pKa = 11.84RR163 pKa = 11.84STAEE167 pKa = 3.86ARR169 pKa = 11.84LKK171 pKa = 9.48TPKK174 pKa = 10.43AKK176 pKa = 10.08NAKK179 pKa = 8.64NPSRR183 pKa = 11.84ALVLARR189 pKa = 11.84RR190 pKa = 11.84LAMSKK195 pKa = 10.04HH196 pKa = 5.51GKK198 pKa = 8.44SANAPTSASSAAVARR213 pKa = 11.84QGNPEE218 pKa = 3.82LSGRR222 pKa = 11.84EE223 pKa = 3.43LAQRR227 pKa = 11.84VRR229 pKa = 11.84EE230 pKa = 4.06LKK232 pKa = 10.61ARR234 pKa = 11.84SGSAGQCVKK243 pKa = 10.91GNNKK247 pKa = 9.46RR248 pKa = 11.84PCGPRR253 pKa = 11.84RR254 pKa = 11.84GMNRR258 pKa = 11.84PKK260 pKa = 11.21DD261 pKa = 3.51NDD263 pKa = 3.62QPVQAADD270 pKa = 3.47AHH272 pKa = 5.91WKK274 pKa = 10.05VGASTTPSGQTVTGTRR290 pKa = 11.84TDD292 pKa = 3.26RR293 pKa = 11.84SVKK296 pKa = 7.61TTGNEE301 pKa = 3.52ASTCRR306 pKa = 11.84TITGTEE312 pKa = 3.88YY313 pKa = 10.03MGADD317 pKa = 3.71VFQSYY322 pKa = 9.69CQSQAPSRR330 pKa = 11.84PRR332 pKa = 11.84KK333 pKa = 9.96VGATPTGYY341 pKa = 11.12GNTVTGTEE349 pKa = 3.87VGRR352 pKa = 11.84SVKK355 pKa = 9.31VTGDD359 pKa = 3.29EE360 pKa = 4.31PGTCKK365 pKa = 10.48SVTGTQYY372 pKa = 11.37LSGNHH377 pKa = 6.21GEE379 pKa = 4.43TWCGSPTQPQPAKK392 pKa = 10.25VGQTRR397 pKa = 11.84TQAEE401 pKa = 4.02QRR403 pKa = 11.84VSGVMVGRR411 pKa = 11.84SARR414 pKa = 11.84VTGDD418 pKa = 3.15EE419 pKa = 4.48PGSGRR424 pKa = 11.84QLTGSQYY431 pKa = 10.47MNPTPDD437 pKa = 3.4MGRR440 pKa = 11.84GPAPAKK446 pKa = 10.14VRR448 pKa = 11.84SFSTLRR454 pKa = 11.84GSAVTGNQVGRR465 pKa = 11.84STKK468 pKa = 9.42VTGDD472 pKa = 3.38EE473 pKa = 4.5PGTCKK478 pKa = 10.52NVTGDD483 pKa = 4.08DD484 pKa = 3.78YY485 pKa = 11.82VGRR488 pKa = 11.84QQYY491 pKa = 10.71DD492 pKa = 3.45AFCEE496 pKa = 4.03SRR498 pKa = 11.84PASEE502 pKa = 3.85AAKK505 pKa = 10.49VGLSATNRR513 pKa = 11.84NQIVSGTYY521 pKa = 8.41TGRR524 pKa = 11.84SPLVTGDD531 pKa = 3.48EE532 pKa = 4.45PGTCKK537 pKa = 10.48VVTGTPYY544 pKa = 10.97AGLEE548 pKa = 3.88QAGNFCSAPEE558 pKa = 3.7QATIVARR565 pKa = 11.84TPAGPGTPGQPMTGLQPGIGGVMTGAEE592 pKa = 4.04RR593 pKa = 11.84GACEE597 pKa = 4.3PVTGTPYY604 pKa = 10.67VGADD608 pKa = 3.45QFLSACDD615 pKa = 3.19QGGAGDD621 pKa = 5.07AVPADD626 pKa = 4.09LPQPLEE632 pKa = 4.08GAPWTRR638 pKa = 11.84FTVMSPFRR646 pKa = 11.84AAHH649 pKa = 5.7TEE651 pKa = 3.58HH652 pKa = 6.82TEE654 pKa = 3.87QKK656 pKa = 10.03RR657 pKa = 11.84RR658 pKa = 11.84LATGVTGTLYY668 pKa = 10.64EE669 pKa = 4.25RR670 pKa = 11.84GGTRR674 pKa = 11.84ITGPFGMAGGRR685 pKa = 11.84VTGTEE690 pKa = 3.89EE691 pKa = 3.94FRR693 pKa = 11.84FDD695 pKa = 3.58QRR697 pKa = 11.84GTNVSLPSRR706 pKa = 11.84NPLQPLASKK715 pKa = 11.07ADD717 pKa = 3.58AFAGATSPAPVAPPAPSRR735 pKa = 11.84VTGEE739 pKa = 3.9GMVQGVRR746 pKa = 11.84ISGDD750 pKa = 2.81DD751 pKa = 2.98WSRR754 pKa = 11.84NEE756 pKa = 4.14RR757 pKa = 11.84VTGTEE762 pKa = 4.09GPSARR767 pKa = 11.84RR768 pKa = 11.84RR769 pKa = 11.84NPSRR773 pKa = 11.84PGPMSAMHH781 pKa = 7.26AFQLKK786 pKa = 9.78RR787 pKa = 11.84NAEE790 pKa = 4.3TEE792 pKa = 4.41VPLSPITGSSGNTDD806 pKa = 2.66KK807 pKa = 11.5GAVVTFSGGARR818 pKa = 11.84GG819 pKa = 3.36
MM1 pKa = 7.15ITTASRR7 pKa = 11.84QSALEE12 pKa = 3.79RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.38ALATAGKK22 pKa = 9.61AAASRR27 pKa = 11.84YY28 pKa = 9.97AKK30 pKa = 9.37TRR32 pKa = 11.84QRR34 pKa = 11.84NISDD38 pKa = 3.46EE39 pKa = 3.87QSPRR43 pKa = 11.84RR44 pKa = 11.84GHH46 pKa = 5.22VVTRR50 pKa = 11.84TVSVTPRR57 pKa = 11.84PRR59 pKa = 11.84PTVVFSPAAVAQPAPAGTPNLNTRR83 pKa = 11.84KK84 pKa = 9.65ISNPTRR90 pKa = 11.84DD91 pKa = 3.05LALARR96 pKa = 11.84RR97 pKa = 11.84QALSRR102 pKa = 11.84QGKK105 pKa = 10.08RR106 pKa = 11.84NDD108 pKa = 3.53PGNNRR113 pKa = 11.84IRR115 pKa = 11.84TEE117 pKa = 3.49VRR119 pKa = 11.84RR120 pKa = 11.84PSRR123 pKa = 11.84TTPQPAVPDD132 pKa = 3.88RR133 pKa = 11.84QDD135 pKa = 3.05TSHH138 pKa = 6.61TCDD141 pKa = 3.01CRR143 pKa = 11.84KK144 pKa = 10.16RR145 pKa = 11.84PGKK148 pKa = 8.41EE149 pKa = 3.43VRR151 pKa = 11.84KK152 pKa = 9.71PNLTSLSTGRR162 pKa = 11.84RR163 pKa = 11.84STAEE167 pKa = 3.86ARR169 pKa = 11.84LKK171 pKa = 9.48TPKK174 pKa = 10.43AKK176 pKa = 10.08NAKK179 pKa = 8.64NPSRR183 pKa = 11.84ALVLARR189 pKa = 11.84RR190 pKa = 11.84LAMSKK195 pKa = 10.04HH196 pKa = 5.51GKK198 pKa = 8.44SANAPTSASSAAVARR213 pKa = 11.84QGNPEE218 pKa = 3.82LSGRR222 pKa = 11.84EE223 pKa = 3.43LAQRR227 pKa = 11.84VRR229 pKa = 11.84EE230 pKa = 4.06LKK232 pKa = 10.61ARR234 pKa = 11.84SGSAGQCVKK243 pKa = 10.91GNNKK247 pKa = 9.46RR248 pKa = 11.84PCGPRR253 pKa = 11.84RR254 pKa = 11.84GMNRR258 pKa = 11.84PKK260 pKa = 11.21DD261 pKa = 3.51NDD263 pKa = 3.62QPVQAADD270 pKa = 3.47AHH272 pKa = 5.91WKK274 pKa = 10.05VGASTTPSGQTVTGTRR290 pKa = 11.84TDD292 pKa = 3.26RR293 pKa = 11.84SVKK296 pKa = 7.61TTGNEE301 pKa = 3.52ASTCRR306 pKa = 11.84TITGTEE312 pKa = 3.88YY313 pKa = 10.03MGADD317 pKa = 3.71VFQSYY322 pKa = 9.69CQSQAPSRR330 pKa = 11.84PRR332 pKa = 11.84KK333 pKa = 9.96VGATPTGYY341 pKa = 11.12GNTVTGTEE349 pKa = 3.87VGRR352 pKa = 11.84SVKK355 pKa = 9.31VTGDD359 pKa = 3.29EE360 pKa = 4.31PGTCKK365 pKa = 10.48SVTGTQYY372 pKa = 11.37LSGNHH377 pKa = 6.21GEE379 pKa = 4.43TWCGSPTQPQPAKK392 pKa = 10.25VGQTRR397 pKa = 11.84TQAEE401 pKa = 4.02QRR403 pKa = 11.84VSGVMVGRR411 pKa = 11.84SARR414 pKa = 11.84VTGDD418 pKa = 3.15EE419 pKa = 4.48PGSGRR424 pKa = 11.84QLTGSQYY431 pKa = 10.47MNPTPDD437 pKa = 3.4MGRR440 pKa = 11.84GPAPAKK446 pKa = 10.14VRR448 pKa = 11.84SFSTLRR454 pKa = 11.84GSAVTGNQVGRR465 pKa = 11.84STKK468 pKa = 9.42VTGDD472 pKa = 3.38EE473 pKa = 4.5PGTCKK478 pKa = 10.52NVTGDD483 pKa = 4.08DD484 pKa = 3.78YY485 pKa = 11.82VGRR488 pKa = 11.84QQYY491 pKa = 10.71DD492 pKa = 3.45AFCEE496 pKa = 4.03SRR498 pKa = 11.84PASEE502 pKa = 3.85AAKK505 pKa = 10.49VGLSATNRR513 pKa = 11.84NQIVSGTYY521 pKa = 8.41TGRR524 pKa = 11.84SPLVTGDD531 pKa = 3.48EE532 pKa = 4.45PGTCKK537 pKa = 10.48VVTGTPYY544 pKa = 10.97AGLEE548 pKa = 3.88QAGNFCSAPEE558 pKa = 3.7QATIVARR565 pKa = 11.84TPAGPGTPGQPMTGLQPGIGGVMTGAEE592 pKa = 4.04RR593 pKa = 11.84GACEE597 pKa = 4.3PVTGTPYY604 pKa = 10.67VGADD608 pKa = 3.45QFLSACDD615 pKa = 3.19QGGAGDD621 pKa = 5.07AVPADD626 pKa = 4.09LPQPLEE632 pKa = 4.08GAPWTRR638 pKa = 11.84FTVMSPFRR646 pKa = 11.84AAHH649 pKa = 5.7TEE651 pKa = 3.58HH652 pKa = 6.82TEE654 pKa = 3.87QKK656 pKa = 10.03RR657 pKa = 11.84RR658 pKa = 11.84LATGVTGTLYY668 pKa = 10.64EE669 pKa = 4.25RR670 pKa = 11.84GGTRR674 pKa = 11.84ITGPFGMAGGRR685 pKa = 11.84VTGTEE690 pKa = 3.89EE691 pKa = 3.94FRR693 pKa = 11.84FDD695 pKa = 3.58QRR697 pKa = 11.84GTNVSLPSRR706 pKa = 11.84NPLQPLASKK715 pKa = 11.07ADD717 pKa = 3.58AFAGATSPAPVAPPAPSRR735 pKa = 11.84VTGEE739 pKa = 3.9GMVQGVRR746 pKa = 11.84ISGDD750 pKa = 2.81DD751 pKa = 2.98WSRR754 pKa = 11.84NEE756 pKa = 4.14RR757 pKa = 11.84VTGTEE762 pKa = 4.09GPSARR767 pKa = 11.84RR768 pKa = 11.84RR769 pKa = 11.84NPSRR773 pKa = 11.84PGPMSAMHH781 pKa = 7.26AFQLKK786 pKa = 9.78RR787 pKa = 11.84NAEE790 pKa = 4.3TEE792 pKa = 4.41VPLSPITGSSGNTDD806 pKa = 2.66KK807 pKa = 11.5GAVVTFSGGARR818 pKa = 11.84GG819 pKa = 3.36
Molecular weight: 86.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
149200 |
21 |
1132 |
280.5 |
30.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.254 ± 0.126 | 1.346 ± 0.044 |
5.22 ± 0.079 | 5.422 ± 0.095 |
3.198 ± 0.067 | 8.252 ± 0.108 |
2.523 ± 0.065 | 4.041 ± 0.087 |
2.902 ± 0.081 | 11.642 ± 0.154 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.121 ± 0.041 | 2.757 ± 0.062 |
5.611 ± 0.084 | 4.703 ± 0.077 |
7.584 ± 0.099 | 5.832 ± 0.079 |
5.41 ± 0.098 | 7.582 ± 0.092 |
1.518 ± 0.049 | 2.082 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
