
Chitinophaga parva
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
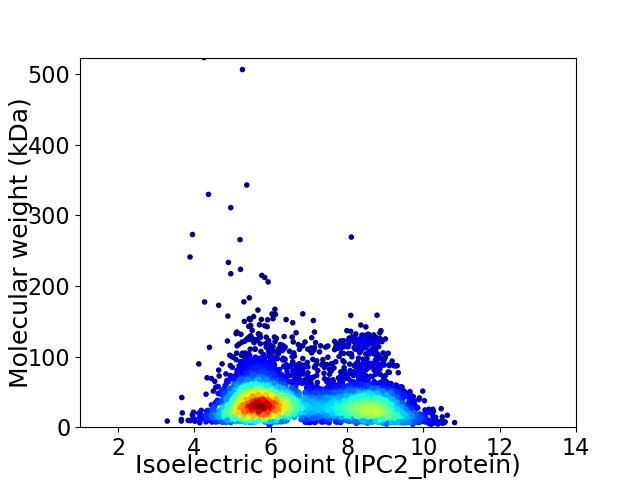
Virtual 2D-PAGE plot for 4954 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
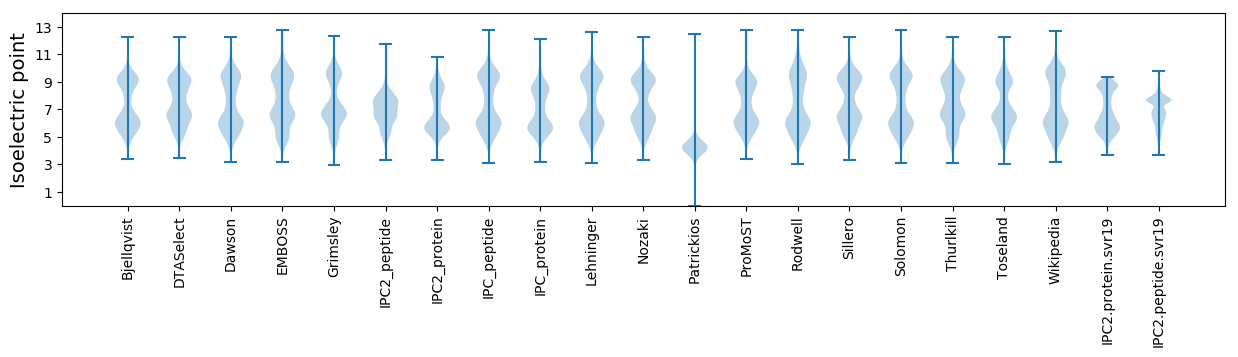
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T7BGG9|A0A2T7BGG9_9BACT Ribulokinase OS=Chitinophaga parva OX=2169414 GN=DCC81_13820 PE=4 SV=1
MM1 pKa = 7.3KK2 pKa = 9.55RR3 pKa = 11.84TFTRR7 pKa = 11.84HH8 pKa = 5.29FPLFLLFMLLSVQHH22 pKa = 5.74VFAVAPYY29 pKa = 8.58PLQRR33 pKa = 11.84WMHH36 pKa = 5.7AVMPHH41 pKa = 6.69DD42 pKa = 3.86VAAPSAKK49 pKa = 9.61PHH51 pKa = 6.6ALLAPATTDD60 pKa = 3.06VSVYY64 pKa = 10.88GDD66 pKa = 3.49NFHH69 pKa = 6.95YY70 pKa = 9.06GTDD73 pKa = 3.02IPLYY77 pKa = 8.58VTFSEE82 pKa = 5.02PINVNLGLGPITLDD96 pKa = 2.86ISMSDD101 pKa = 3.08GTSRR105 pKa = 11.84TFVCVGRR112 pKa = 11.84DD113 pKa = 3.39PADD116 pKa = 3.47NTSLEE121 pKa = 3.94MHH123 pKa = 6.1YY124 pKa = 9.51TVQDD128 pKa = 2.89GDD130 pKa = 4.02FQDD133 pKa = 4.1SGNLPFAAAVVAPAGAITSLADD155 pKa = 3.43GTPVDD160 pKa = 5.02LSTAGATDD168 pKa = 3.74DD169 pKa = 3.19TWGNFVSGVVPHH181 pKa = 6.5VVLTPDD187 pKa = 4.03PLPAKK192 pKa = 10.39VSGTFTVTATFSPEE206 pKa = 3.61SMIGVTQSIFTATNATLSAFQDD228 pKa = 3.77NVDD231 pKa = 3.76GQHH234 pKa = 6.08KK235 pKa = 10.1VFAISVTPIADD246 pKa = 3.7GPLTVSIPANMAANIQSNYY265 pKa = 9.76NFASNVLTATVDD277 pKa = 3.31QTAPSITSVDD287 pKa = 3.86VPPDD291 pKa = 3.48DD292 pKa = 4.87YY293 pKa = 12.14YY294 pKa = 11.94NATNTLQFTVHH305 pKa = 6.09FSEE308 pKa = 4.35NVFVTGTPVLYY319 pKa = 10.69LNIGGTPVQATYY331 pKa = 10.5TSGTGSSALLFSYY344 pKa = 10.45HH345 pKa = 7.03ILDD348 pKa = 4.14GQADD352 pKa = 3.96ADD354 pKa = 4.71GITIDD359 pKa = 4.02SLGLNGSATIGDD371 pKa = 3.95DD372 pKa = 3.32THH374 pKa = 7.58NNANLTLNGVPSTAGVKK391 pKa = 10.49VNTTHH396 pKa = 6.54PTVTLTTAAPAVVNGPFNVTATFSEE421 pKa = 4.79WVSGLDD427 pKa = 3.27AFDD430 pKa = 3.54FTIVNGSVNAPTTSDD445 pKa = 3.31NITYY449 pKa = 8.0TFQVVPATPDD459 pKa = 3.19QPVSITLPADD469 pKa = 3.02KK470 pKa = 10.92AEE472 pKa = 4.62NIGHH476 pKa = 6.52NGNLASNTISLQFDD490 pKa = 3.61PTAPAITVIEE500 pKa = 4.85GPANATYY507 pKa = 10.39IAGQDD512 pKa = 3.37LTFKK516 pKa = 10.84VVFSEE521 pKa = 4.26KK522 pKa = 10.66VNVTGVPYY530 pKa = 10.91LDD532 pKa = 4.62LNIGGTTVQAQYY544 pKa = 11.39QSGTGDD550 pKa = 3.03SVLYY554 pKa = 9.63FKK556 pKa = 10.5YY557 pKa = 9.24TVVDD561 pKa = 4.33GNHH564 pKa = 6.8DD565 pKa = 3.66ADD567 pKa = 5.17GIGVTAANAGTGTIKK582 pKa = 10.85DD583 pKa = 3.49IATNDD588 pKa = 3.24ADD590 pKa = 4.09LTINPAVVADD600 pKa = 3.87LTGVLVNTQHH610 pKa = 6.98PAVTITGPSTSASSVNVTITFSEE633 pKa = 4.09AMGAVDD639 pKa = 4.0AGKK642 pKa = 10.19FILGGTVTGTSLTGLTTLDD661 pKa = 3.77NITYY665 pKa = 8.94TGTLNFPANVTGTVTLSLPADD686 pKa = 3.75VVKK689 pKa = 10.89SAVGNNNQASNVYY702 pKa = 8.44TVNVDD707 pKa = 3.07NAGPVVTAVSGPADD721 pKa = 3.12KK722 pKa = 10.31TYY724 pKa = 10.95RR725 pKa = 11.84QGEE728 pKa = 4.14QLQFTVTFNEE738 pKa = 4.52PVTVDD743 pKa = 3.31QTGGTPQLPIIIGSSTVWADD763 pKa = 3.52YY764 pKa = 11.32VSGTGTTQLVFSYY777 pKa = 10.17TVLPNQYY784 pKa = 10.22DD785 pKa = 3.71ADD787 pKa = 4.11GVTTGTDD794 pKa = 3.11LAANGGTLQDD804 pKa = 3.97ALGNDD809 pKa = 3.92ASLTLHH815 pKa = 6.21NVAATTGVLVNARR828 pKa = 11.84IPSVTLGPVAPGVTGPFTVTATFSEE853 pKa = 4.85KK854 pKa = 9.3VTGFDD859 pKa = 3.31ATDD862 pKa = 3.24VTLVNGTAGTPVSSDD877 pKa = 2.68GGKK880 pKa = 8.4TYY882 pKa = 10.42TFTVTPAADD891 pKa = 3.55GTVSVTIPANAAFNTGNNGNTASNTITTTADD922 pKa = 2.8VTAPYY927 pKa = 8.19ITSVDD932 pKa = 3.45VPAPLVYY939 pKa = 10.35NSSMVLTFQVHH950 pKa = 5.44FSEE953 pKa = 4.46NAVVTNVPGLPIDD966 pKa = 3.3IGGQTRR972 pKa = 11.84EE973 pKa = 4.08AIYY976 pKa = 10.14TGGTGTDD983 pKa = 3.04TWSFAYY989 pKa = 10.14LVQDD993 pKa = 4.36GDD995 pKa = 3.32QDD997 pKa = 3.36MDD999 pKa = 4.6GITLGTALDD1008 pKa = 3.59ISTSIIRR1015 pKa = 11.84DD1016 pKa = 3.32AAGNDD1021 pKa = 3.4ADD1023 pKa = 4.1VTLHH1027 pKa = 6.22NVGNTTGVKK1036 pKa = 10.37VNTAHH1041 pKa = 6.51PTVTLSGAPASAHH1054 pKa = 5.24TPFTITATFSEE1065 pKa = 5.41VVTGLAVTDD1074 pKa = 5.17FSAPNGATFSNLQSADD1090 pKa = 3.59GITYY1094 pKa = 8.43TVLVTPTADD1103 pKa = 3.24GTLTISLPANIAVNVGNNGNLASNTLSSSVDD1134 pKa = 3.24VSAPAVTSVDD1144 pKa = 3.38VPANATYY1151 pKa = 8.44KK1152 pKa = 9.98TGQVLNFTVHH1162 pKa = 6.05FSEE1165 pKa = 4.7VVNVTGNPALPITIGTQTVLAAYY1188 pKa = 8.18TGGAGTTALTFAYY1201 pKa = 8.59TILDD1205 pKa = 3.58GQMDD1209 pKa = 5.05LDD1211 pKa = 5.14GIALAASLLPNGATIQDD1228 pKa = 3.63AATNNANLTLNNVAATNNVLVNTSHH1253 pKa = 6.41PTVVITGPATASGTFTATVTFSAVMTGVTLSDD1285 pKa = 3.57FAVTNGTLSNLVTTDD1300 pKa = 3.24DD1301 pKa = 3.13VTYY1304 pKa = 9.92TVDD1307 pKa = 3.24VTPAADD1313 pKa = 3.82GSLTLMLPADD1323 pKa = 3.82VAQNSIGSGNQASNTLSVNITLPVPEE1349 pKa = 4.27ITQVDD1354 pKa = 4.27VPVNGTYY1361 pKa = 7.32TTGQDD1366 pKa = 3.18LTFTVHH1372 pKa = 6.42FSEE1375 pKa = 4.71VVNVTGTPALPVTIGTQTVLATYY1398 pKa = 10.74VSGTGSDD1405 pKa = 3.08ILTFSYY1411 pKa = 10.33TVLDD1415 pKa = 3.8GQMDD1419 pKa = 4.81LDD1421 pKa = 5.09GISLATSLLPNGATIRR1437 pKa = 11.84DD1438 pKa = 3.58AAARR1442 pKa = 11.84DD1443 pKa = 3.55ADD1445 pKa = 4.02FTLHH1449 pKa = 6.17NVASTTNVLVNTHH1462 pKa = 6.96LPTVVISGPATISSSFTATITFSEE1486 pKa = 4.45AMTGLTLSDD1495 pKa = 3.75FLVSNATLTNLTTTDD1510 pKa = 4.19GIVYY1514 pKa = 8.62TVDD1517 pKa = 3.2VTPVADD1523 pKa = 4.76GSLTLQLPANVAVNTQAAGNKK1544 pKa = 9.19ASNILTVTADD1554 pKa = 3.61LAVPVITQVDD1564 pKa = 4.27VPADD1568 pKa = 3.66NIYY1571 pKa = 11.01NAGSVLTFTVHH1582 pKa = 5.6YY1583 pKa = 9.15SEE1585 pKa = 5.05NVAITGTPKK1594 pKa = 10.67LPIIIGTQPVQASYY1608 pKa = 11.72VSGSGTNLLVFTYY1621 pKa = 9.21TVQNGDD1627 pKa = 3.67LDD1629 pKa = 3.69MDD1631 pKa = 5.2GIALGSALILNGGSIKK1647 pKa = 10.75DD1648 pKa = 3.92GISNDD1653 pKa = 3.85GVLTLNNIASTANVLVNTAHH1673 pKa = 6.07PTVVISSPAPTVTAPFSITITFSEE1697 pKa = 4.61AVTGFTSSDD1706 pKa = 3.34FVLTNATVSNLQTSDD1721 pKa = 3.1NKK1723 pKa = 10.43VYY1725 pKa = 9.29TALVIPGAAGTLSIDD1740 pKa = 3.27IPADD1744 pKa = 3.27VAMNAANNGNTASNTVSTMVDD1765 pKa = 3.97FNAPYY1770 pKa = 8.18ITQVNVPADD1779 pKa = 3.79NTYY1782 pKa = 10.88HH1783 pKa = 6.66SGDD1786 pKa = 3.53VLSFSVTFNKK1796 pKa = 9.99GVYY1799 pKa = 8.16VTGTPSLDD1807 pKa = 3.3IIIGAKK1813 pKa = 8.01TAQATLVSGSGTGVLTFNYY1832 pKa = 9.21TVVDD1836 pKa = 3.73GDD1838 pKa = 3.68YY1839 pKa = 11.56DD1840 pKa = 3.44MDD1842 pKa = 5.46GIALTTSLNLNGGTITDD1859 pKa = 3.88ANSNNADD1866 pKa = 3.46VTVKK1870 pKa = 10.59NAASTSNIFVNAIRR1884 pKa = 11.84PSVVLTGTGTGSAPFTITITFSEE1907 pKa = 4.75AVTGLTAADD1916 pKa = 3.74FAIVNATGSNLQTTDD1931 pKa = 2.85NKK1933 pKa = 10.88TYY1935 pKa = 10.01TLLITPGGGGTITINLPANMAQNAGGNGNTASNTLSISVGSPAPVINTPPAFQAVEE1991 pKa = 3.97HH1992 pKa = 6.73SPVGTAVGTLTATASTGTLQQWTITSDD2019 pKa = 3.14NSGGAFQIDD2028 pKa = 4.67PNTGEE2033 pKa = 3.99ITVKK2037 pKa = 10.74DD2038 pKa = 3.88VTLLDD2043 pKa = 3.61AKK2045 pKa = 10.81VYY2047 pKa = 9.4STVTLTVTVSDD2058 pKa = 4.55GFNTSAPATVHH2069 pKa = 6.42ISVTPAPAAVNQAPILDD2086 pKa = 4.0AVTDD2090 pKa = 3.94KK2091 pKa = 9.83QTCAVPAKK2099 pKa = 10.42QSIQLTGGSPVEE2111 pKa = 4.43AGQQLTYY2118 pKa = 10.21TISADD2123 pKa = 3.27KK2124 pKa = 11.25DD2125 pKa = 3.53FFTALTINNSGLISYY2140 pKa = 7.41TLKK2143 pKa = 10.75PNTTGKK2149 pKa = 10.75VNFTVVLKK2157 pKa = 10.82DD2158 pKa = 4.42DD2159 pKa = 4.26GGTANGGVDD2168 pKa = 3.52SLRR2171 pKa = 11.84VTFALTINALPDD2183 pKa = 3.33VSISADD2189 pKa = 3.22KK2190 pKa = 10.87EE2191 pKa = 4.24GEE2193 pKa = 4.08INQGEE2198 pKa = 4.96SLTLTADD2205 pKa = 3.3APGASQYY2212 pKa = 11.06AWNTGAEE2219 pKa = 4.21GRR2221 pKa = 11.84QLNVQPSEE2229 pKa = 3.92DD2230 pKa = 3.4TTYY2233 pKa = 10.66MVTASTGNGCTSSASYY2249 pKa = 10.35NVRR2252 pKa = 11.84VKK2254 pKa = 10.16PAPDD2258 pKa = 3.43TKK2260 pKa = 9.95ITATNFLTPNGDD2272 pKa = 3.07GRR2274 pKa = 11.84NDD2276 pKa = 2.93RR2277 pKa = 11.84WIVTNLQNYY2286 pKa = 6.06GTYY2289 pKa = 10.31DD2290 pKa = 2.9ITIFDD2295 pKa = 3.58RR2296 pKa = 11.84AGRR2299 pKa = 11.84IVYY2302 pKa = 7.51YY2303 pKa = 9.63TRR2305 pKa = 11.84NYY2307 pKa = 9.16NNDD2310 pKa = 2.91WGGTANGDD2318 pKa = 3.69PLAEE2322 pKa = 3.82GTYY2325 pKa = 9.81YY2326 pKa = 10.94YY2327 pKa = 10.4VINVQGQQPVKK2338 pKa = 10.59GFVTIVRR2345 pKa = 11.84DD2346 pKa = 3.51IKK2348 pKa = 10.92KK2349 pKa = 10.48
MM1 pKa = 7.3KK2 pKa = 9.55RR3 pKa = 11.84TFTRR7 pKa = 11.84HH8 pKa = 5.29FPLFLLFMLLSVQHH22 pKa = 5.74VFAVAPYY29 pKa = 8.58PLQRR33 pKa = 11.84WMHH36 pKa = 5.7AVMPHH41 pKa = 6.69DD42 pKa = 3.86VAAPSAKK49 pKa = 9.61PHH51 pKa = 6.6ALLAPATTDD60 pKa = 3.06VSVYY64 pKa = 10.88GDD66 pKa = 3.49NFHH69 pKa = 6.95YY70 pKa = 9.06GTDD73 pKa = 3.02IPLYY77 pKa = 8.58VTFSEE82 pKa = 5.02PINVNLGLGPITLDD96 pKa = 2.86ISMSDD101 pKa = 3.08GTSRR105 pKa = 11.84TFVCVGRR112 pKa = 11.84DD113 pKa = 3.39PADD116 pKa = 3.47NTSLEE121 pKa = 3.94MHH123 pKa = 6.1YY124 pKa = 9.51TVQDD128 pKa = 2.89GDD130 pKa = 4.02FQDD133 pKa = 4.1SGNLPFAAAVVAPAGAITSLADD155 pKa = 3.43GTPVDD160 pKa = 5.02LSTAGATDD168 pKa = 3.74DD169 pKa = 3.19TWGNFVSGVVPHH181 pKa = 6.5VVLTPDD187 pKa = 4.03PLPAKK192 pKa = 10.39VSGTFTVTATFSPEE206 pKa = 3.61SMIGVTQSIFTATNATLSAFQDD228 pKa = 3.77NVDD231 pKa = 3.76GQHH234 pKa = 6.08KK235 pKa = 10.1VFAISVTPIADD246 pKa = 3.7GPLTVSIPANMAANIQSNYY265 pKa = 9.76NFASNVLTATVDD277 pKa = 3.31QTAPSITSVDD287 pKa = 3.86VPPDD291 pKa = 3.48DD292 pKa = 4.87YY293 pKa = 12.14YY294 pKa = 11.94NATNTLQFTVHH305 pKa = 6.09FSEE308 pKa = 4.35NVFVTGTPVLYY319 pKa = 10.69LNIGGTPVQATYY331 pKa = 10.5TSGTGSSALLFSYY344 pKa = 10.45HH345 pKa = 7.03ILDD348 pKa = 4.14GQADD352 pKa = 3.96ADD354 pKa = 4.71GITIDD359 pKa = 4.02SLGLNGSATIGDD371 pKa = 3.95DD372 pKa = 3.32THH374 pKa = 7.58NNANLTLNGVPSTAGVKK391 pKa = 10.49VNTTHH396 pKa = 6.54PTVTLTTAAPAVVNGPFNVTATFSEE421 pKa = 4.79WVSGLDD427 pKa = 3.27AFDD430 pKa = 3.54FTIVNGSVNAPTTSDD445 pKa = 3.31NITYY449 pKa = 8.0TFQVVPATPDD459 pKa = 3.19QPVSITLPADD469 pKa = 3.02KK470 pKa = 10.92AEE472 pKa = 4.62NIGHH476 pKa = 6.52NGNLASNTISLQFDD490 pKa = 3.61PTAPAITVIEE500 pKa = 4.85GPANATYY507 pKa = 10.39IAGQDD512 pKa = 3.37LTFKK516 pKa = 10.84VVFSEE521 pKa = 4.26KK522 pKa = 10.66VNVTGVPYY530 pKa = 10.91LDD532 pKa = 4.62LNIGGTTVQAQYY544 pKa = 11.39QSGTGDD550 pKa = 3.03SVLYY554 pKa = 9.63FKK556 pKa = 10.5YY557 pKa = 9.24TVVDD561 pKa = 4.33GNHH564 pKa = 6.8DD565 pKa = 3.66ADD567 pKa = 5.17GIGVTAANAGTGTIKK582 pKa = 10.85DD583 pKa = 3.49IATNDD588 pKa = 3.24ADD590 pKa = 4.09LTINPAVVADD600 pKa = 3.87LTGVLVNTQHH610 pKa = 6.98PAVTITGPSTSASSVNVTITFSEE633 pKa = 4.09AMGAVDD639 pKa = 4.0AGKK642 pKa = 10.19FILGGTVTGTSLTGLTTLDD661 pKa = 3.77NITYY665 pKa = 8.94TGTLNFPANVTGTVTLSLPADD686 pKa = 3.75VVKK689 pKa = 10.89SAVGNNNQASNVYY702 pKa = 8.44TVNVDD707 pKa = 3.07NAGPVVTAVSGPADD721 pKa = 3.12KK722 pKa = 10.31TYY724 pKa = 10.95RR725 pKa = 11.84QGEE728 pKa = 4.14QLQFTVTFNEE738 pKa = 4.52PVTVDD743 pKa = 3.31QTGGTPQLPIIIGSSTVWADD763 pKa = 3.52YY764 pKa = 11.32VSGTGTTQLVFSYY777 pKa = 10.17TVLPNQYY784 pKa = 10.22DD785 pKa = 3.71ADD787 pKa = 4.11GVTTGTDD794 pKa = 3.11LAANGGTLQDD804 pKa = 3.97ALGNDD809 pKa = 3.92ASLTLHH815 pKa = 6.21NVAATTGVLVNARR828 pKa = 11.84IPSVTLGPVAPGVTGPFTVTATFSEE853 pKa = 4.85KK854 pKa = 9.3VTGFDD859 pKa = 3.31ATDD862 pKa = 3.24VTLVNGTAGTPVSSDD877 pKa = 2.68GGKK880 pKa = 8.4TYY882 pKa = 10.42TFTVTPAADD891 pKa = 3.55GTVSVTIPANAAFNTGNNGNTASNTITTTADD922 pKa = 2.8VTAPYY927 pKa = 8.19ITSVDD932 pKa = 3.45VPAPLVYY939 pKa = 10.35NSSMVLTFQVHH950 pKa = 5.44FSEE953 pKa = 4.46NAVVTNVPGLPIDD966 pKa = 3.3IGGQTRR972 pKa = 11.84EE973 pKa = 4.08AIYY976 pKa = 10.14TGGTGTDD983 pKa = 3.04TWSFAYY989 pKa = 10.14LVQDD993 pKa = 4.36GDD995 pKa = 3.32QDD997 pKa = 3.36MDD999 pKa = 4.6GITLGTALDD1008 pKa = 3.59ISTSIIRR1015 pKa = 11.84DD1016 pKa = 3.32AAGNDD1021 pKa = 3.4ADD1023 pKa = 4.1VTLHH1027 pKa = 6.22NVGNTTGVKK1036 pKa = 10.37VNTAHH1041 pKa = 6.51PTVTLSGAPASAHH1054 pKa = 5.24TPFTITATFSEE1065 pKa = 5.41VVTGLAVTDD1074 pKa = 5.17FSAPNGATFSNLQSADD1090 pKa = 3.59GITYY1094 pKa = 8.43TVLVTPTADD1103 pKa = 3.24GTLTISLPANIAVNVGNNGNLASNTLSSSVDD1134 pKa = 3.24VSAPAVTSVDD1144 pKa = 3.38VPANATYY1151 pKa = 8.44KK1152 pKa = 9.98TGQVLNFTVHH1162 pKa = 6.05FSEE1165 pKa = 4.7VVNVTGNPALPITIGTQTVLAAYY1188 pKa = 8.18TGGAGTTALTFAYY1201 pKa = 8.59TILDD1205 pKa = 3.58GQMDD1209 pKa = 5.05LDD1211 pKa = 5.14GIALAASLLPNGATIQDD1228 pKa = 3.63AATNNANLTLNNVAATNNVLVNTSHH1253 pKa = 6.41PTVVITGPATASGTFTATVTFSAVMTGVTLSDD1285 pKa = 3.57FAVTNGTLSNLVTTDD1300 pKa = 3.24DD1301 pKa = 3.13VTYY1304 pKa = 9.92TVDD1307 pKa = 3.24VTPAADD1313 pKa = 3.82GSLTLMLPADD1323 pKa = 3.82VAQNSIGSGNQASNTLSVNITLPVPEE1349 pKa = 4.27ITQVDD1354 pKa = 4.27VPVNGTYY1361 pKa = 7.32TTGQDD1366 pKa = 3.18LTFTVHH1372 pKa = 6.42FSEE1375 pKa = 4.71VVNVTGTPALPVTIGTQTVLATYY1398 pKa = 10.74VSGTGSDD1405 pKa = 3.08ILTFSYY1411 pKa = 10.33TVLDD1415 pKa = 3.8GQMDD1419 pKa = 4.81LDD1421 pKa = 5.09GISLATSLLPNGATIRR1437 pKa = 11.84DD1438 pKa = 3.58AAARR1442 pKa = 11.84DD1443 pKa = 3.55ADD1445 pKa = 4.02FTLHH1449 pKa = 6.17NVASTTNVLVNTHH1462 pKa = 6.96LPTVVISGPATISSSFTATITFSEE1486 pKa = 4.45AMTGLTLSDD1495 pKa = 3.75FLVSNATLTNLTTTDD1510 pKa = 4.19GIVYY1514 pKa = 8.62TVDD1517 pKa = 3.2VTPVADD1523 pKa = 4.76GSLTLQLPANVAVNTQAAGNKK1544 pKa = 9.19ASNILTVTADD1554 pKa = 3.61LAVPVITQVDD1564 pKa = 4.27VPADD1568 pKa = 3.66NIYY1571 pKa = 11.01NAGSVLTFTVHH1582 pKa = 5.6YY1583 pKa = 9.15SEE1585 pKa = 5.05NVAITGTPKK1594 pKa = 10.67LPIIIGTQPVQASYY1608 pKa = 11.72VSGSGTNLLVFTYY1621 pKa = 9.21TVQNGDD1627 pKa = 3.67LDD1629 pKa = 3.69MDD1631 pKa = 5.2GIALGSALILNGGSIKK1647 pKa = 10.75DD1648 pKa = 3.92GISNDD1653 pKa = 3.85GVLTLNNIASTANVLVNTAHH1673 pKa = 6.07PTVVISSPAPTVTAPFSITITFSEE1697 pKa = 4.61AVTGFTSSDD1706 pKa = 3.34FVLTNATVSNLQTSDD1721 pKa = 3.1NKK1723 pKa = 10.43VYY1725 pKa = 9.29TALVIPGAAGTLSIDD1740 pKa = 3.27IPADD1744 pKa = 3.27VAMNAANNGNTASNTVSTMVDD1765 pKa = 3.97FNAPYY1770 pKa = 8.18ITQVNVPADD1779 pKa = 3.79NTYY1782 pKa = 10.88HH1783 pKa = 6.66SGDD1786 pKa = 3.53VLSFSVTFNKK1796 pKa = 9.99GVYY1799 pKa = 8.16VTGTPSLDD1807 pKa = 3.3IIIGAKK1813 pKa = 8.01TAQATLVSGSGTGVLTFNYY1832 pKa = 9.21TVVDD1836 pKa = 3.73GDD1838 pKa = 3.68YY1839 pKa = 11.56DD1840 pKa = 3.44MDD1842 pKa = 5.46GIALTTSLNLNGGTITDD1859 pKa = 3.88ANSNNADD1866 pKa = 3.46VTVKK1870 pKa = 10.59NAASTSNIFVNAIRR1884 pKa = 11.84PSVVLTGTGTGSAPFTITITFSEE1907 pKa = 4.75AVTGLTAADD1916 pKa = 3.74FAIVNATGSNLQTTDD1931 pKa = 2.85NKK1933 pKa = 10.88TYY1935 pKa = 10.01TLLITPGGGGTITINLPANMAQNAGGNGNTASNTLSISVGSPAPVINTPPAFQAVEE1991 pKa = 3.97HH1992 pKa = 6.73SPVGTAVGTLTATASTGTLQQWTITSDD2019 pKa = 3.14NSGGAFQIDD2028 pKa = 4.67PNTGEE2033 pKa = 3.99ITVKK2037 pKa = 10.74DD2038 pKa = 3.88VTLLDD2043 pKa = 3.61AKK2045 pKa = 10.81VYY2047 pKa = 9.4STVTLTVTVSDD2058 pKa = 4.55GFNTSAPATVHH2069 pKa = 6.42ISVTPAPAAVNQAPILDD2086 pKa = 4.0AVTDD2090 pKa = 3.94KK2091 pKa = 9.83QTCAVPAKK2099 pKa = 10.42QSIQLTGGSPVEE2111 pKa = 4.43AGQQLTYY2118 pKa = 10.21TISADD2123 pKa = 3.27KK2124 pKa = 11.25DD2125 pKa = 3.53FFTALTINNSGLISYY2140 pKa = 7.41TLKK2143 pKa = 10.75PNTTGKK2149 pKa = 10.75VNFTVVLKK2157 pKa = 10.82DD2158 pKa = 4.42DD2159 pKa = 4.26GGTANGGVDD2168 pKa = 3.52SLRR2171 pKa = 11.84VTFALTINALPDD2183 pKa = 3.33VSISADD2189 pKa = 3.22KK2190 pKa = 10.87EE2191 pKa = 4.24GEE2193 pKa = 4.08INQGEE2198 pKa = 4.96SLTLTADD2205 pKa = 3.3APGASQYY2212 pKa = 11.06AWNTGAEE2219 pKa = 4.21GRR2221 pKa = 11.84QLNVQPSEE2229 pKa = 3.92DD2230 pKa = 3.4TTYY2233 pKa = 10.66MVTASTGNGCTSSASYY2249 pKa = 10.35NVRR2252 pKa = 11.84VKK2254 pKa = 10.16PAPDD2258 pKa = 3.43TKK2260 pKa = 9.95ITATNFLTPNGDD2272 pKa = 3.07GRR2274 pKa = 11.84NDD2276 pKa = 2.93RR2277 pKa = 11.84WIVTNLQNYY2286 pKa = 6.06GTYY2289 pKa = 10.31DD2290 pKa = 2.9ITIFDD2295 pKa = 3.58RR2296 pKa = 11.84AGRR2299 pKa = 11.84IVYY2302 pKa = 7.51YY2303 pKa = 9.63TRR2305 pKa = 11.84NYY2307 pKa = 9.16NNDD2310 pKa = 2.91WGGTANGDD2318 pKa = 3.69PLAEE2322 pKa = 3.82GTYY2325 pKa = 9.81YY2326 pKa = 10.94YY2327 pKa = 10.4VINVQGQQPVKK2338 pKa = 10.59GFVTIVRR2345 pKa = 11.84DD2346 pKa = 3.51IKK2348 pKa = 10.92KK2349 pKa = 10.48
Molecular weight: 241.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T7BN60|A0A2T7BN60_9BACT Histidine kinase OS=Chitinophaga parva OX=2169414 GN=DCC81_06375 PE=4 SV=1
MM1 pKa = 7.23PHH3 pKa = 6.39FMKK6 pKa = 10.32IVYY9 pKa = 10.0RR10 pKa = 11.84IILAVLGLFGIFLCGTGVYY29 pKa = 9.88LALKK33 pKa = 10.47AIGQGQLAAGNKK45 pKa = 9.05HH46 pKa = 5.73RR47 pKa = 11.84VLTIITCIQPFAMLLVAICAALALRR72 pKa = 11.84RR73 pKa = 11.84EE74 pKa = 4.94LKK76 pKa = 9.22TAPHH80 pKa = 7.16PPRR83 pKa = 5.61
MM1 pKa = 7.23PHH3 pKa = 6.39FMKK6 pKa = 10.32IVYY9 pKa = 10.0RR10 pKa = 11.84IILAVLGLFGIFLCGTGVYY29 pKa = 9.88LALKK33 pKa = 10.47AIGQGQLAAGNKK45 pKa = 9.05HH46 pKa = 5.73RR47 pKa = 11.84VLTIITCIQPFAMLLVAICAALALRR72 pKa = 11.84RR73 pKa = 11.84EE74 pKa = 4.94LKK76 pKa = 9.22TAPHH80 pKa = 7.16PPRR83 pKa = 5.61
Molecular weight: 9.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1802866 |
26 |
5109 |
363.9 |
40.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.06 ± 0.039 | 0.831 ± 0.014 |
5.332 ± 0.022 | 4.956 ± 0.038 |
4.494 ± 0.024 | 7.254 ± 0.033 |
2.299 ± 0.018 | 5.841 ± 0.024 |
5.436 ± 0.034 | 9.716 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.423 ± 0.016 | 4.853 ± 0.03 |
4.41 ± 0.021 | 4.357 ± 0.022 |
4.681 ± 0.025 | 5.723 ± 0.027 |
6.133 ± 0.051 | 6.77 ± 0.032 |
1.307 ± 0.012 | 4.127 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
