
Pseudoalteromonas rubra
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
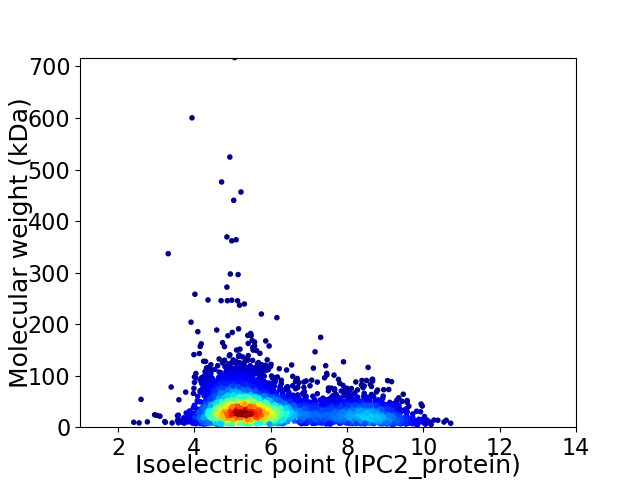
Virtual 2D-PAGE plot for 4390 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F4QPL9|A0A0F4QPL9_9GAMM Bacterioferritin OS=Pseudoalteromonas rubra OX=43658 GN=bfr PE=3 SV=1
MM1 pKa = 8.15DD2 pKa = 4.58YY3 pKa = 9.86STSDD7 pKa = 3.47LCDD10 pKa = 3.4HH11 pKa = 6.92FADD14 pKa = 4.11AVDD17 pKa = 3.54VLEE20 pKa = 4.59PMFINFGGQSAFSGRR35 pKa = 11.84IKK37 pKa = 9.73TLKK40 pKa = 10.25CFEE43 pKa = 4.42NNEE46 pKa = 4.71LINQVLQQDD55 pKa = 3.9GTGQVLLVDD64 pKa = 4.63GGGSTRR70 pKa = 11.84RR71 pKa = 11.84ALIDD75 pKa = 3.32IEE77 pKa = 4.1LAEE80 pKa = 4.94LAIEE84 pKa = 4.36NNWQGIVVYY93 pKa = 10.01GAVRR97 pKa = 11.84HH98 pKa = 4.91VEE100 pKa = 3.96EE101 pKa = 6.22LEE103 pKa = 4.05EE104 pKa = 5.45CDD106 pKa = 5.7LGVQAIASIPVAADD120 pKa = 3.06SHH122 pKa = 6.74GSGEE126 pKa = 3.9QGIPVNFAGVSFYY139 pKa = 11.31DD140 pKa = 4.49DD141 pKa = 4.15DD142 pKa = 4.23YY143 pKa = 11.77LYY145 pKa = 11.4ADD147 pKa = 3.58ATGIIISAEE156 pKa = 3.96EE157 pKa = 3.96LLLEE161 pKa = 4.43VDD163 pKa = 4.69DD164 pKa = 4.83EE165 pKa = 4.36QGEE168 pKa = 4.24
MM1 pKa = 8.15DD2 pKa = 4.58YY3 pKa = 9.86STSDD7 pKa = 3.47LCDD10 pKa = 3.4HH11 pKa = 6.92FADD14 pKa = 4.11AVDD17 pKa = 3.54VLEE20 pKa = 4.59PMFINFGGQSAFSGRR35 pKa = 11.84IKK37 pKa = 9.73TLKK40 pKa = 10.25CFEE43 pKa = 4.42NNEE46 pKa = 4.71LINQVLQQDD55 pKa = 3.9GTGQVLLVDD64 pKa = 4.63GGGSTRR70 pKa = 11.84RR71 pKa = 11.84ALIDD75 pKa = 3.32IEE77 pKa = 4.1LAEE80 pKa = 4.94LAIEE84 pKa = 4.36NNWQGIVVYY93 pKa = 10.01GAVRR97 pKa = 11.84HH98 pKa = 4.91VEE100 pKa = 3.96EE101 pKa = 6.22LEE103 pKa = 4.05EE104 pKa = 5.45CDD106 pKa = 5.7LGVQAIASIPVAADD120 pKa = 3.06SHH122 pKa = 6.74GSGEE126 pKa = 3.9QGIPVNFAGVSFYY139 pKa = 11.31DD140 pKa = 4.49DD141 pKa = 4.15DD142 pKa = 4.23YY143 pKa = 11.77LYY145 pKa = 11.4ADD147 pKa = 3.58ATGIIISAEE156 pKa = 3.96EE157 pKa = 3.96LLLEE161 pKa = 4.43VDD163 pKa = 4.69DD164 pKa = 4.83EE165 pKa = 4.36QGEE168 pKa = 4.24
Molecular weight: 18.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4QL14|A0A0F4QL14_9GAMM Uncharacterized protein OS=Pseudoalteromonas rubra OX=43658 GN=TW77_13870 PE=4 SV=1
MM1 pKa = 7.57GSDD4 pKa = 3.52NSPALSLGEE13 pKa = 3.94PLQVCDD19 pKa = 4.09KK20 pKa = 9.45QRR22 pKa = 11.84KK23 pKa = 7.32RR24 pKa = 11.84AVRR27 pKa = 11.84KK28 pKa = 9.07RR29 pKa = 11.84WILTGSTSAVLKK41 pKa = 10.71QLTLPMLWAILALFSADD58 pKa = 4.51LIEE61 pKa = 5.65LYY63 pKa = 10.22FASRR67 pKa = 11.84LGVEE71 pKa = 4.71EE72 pKa = 4.12LTAMSFTLPVQATLFAFAIGLGIVVATRR100 pKa = 11.84LTQAKK105 pKa = 9.27EE106 pKa = 4.14VEE108 pKa = 4.12QLAAVSLIFTVLVGATLAAVIWLGLQPLLTLLGFAEE144 pKa = 5.1FAARR148 pKa = 11.84EE149 pKa = 4.25QVWPTLEE156 pKa = 5.32RR157 pKa = 11.84YY158 pKa = 6.77MQYY161 pKa = 11.22RR162 pKa = 11.84LGAVVFFFIVMVVFGVLRR180 pKa = 11.84AFGNMRR186 pKa = 11.84GAAQLLVTFAGLQIVISAALFTPWAHH212 pKa = 6.27SALPLSGLEE221 pKa = 4.05RR222 pKa = 11.84LGVAHH227 pKa = 7.12FIAAVSASLYY237 pKa = 10.92ALYY240 pKa = 10.83LLRR243 pKa = 11.84IKK245 pKa = 10.68EE246 pKa = 4.19NISLKK251 pKa = 10.33VRR253 pKa = 11.84LLSQRR258 pKa = 11.84SRR260 pKa = 11.84QAFRR264 pKa = 11.84RR265 pKa = 11.84LFRR268 pKa = 11.84LFVPVVAMQLLTPLAQSLLMMIVASQGSDD297 pKa = 2.62AVAAFGVVMRR307 pKa = 11.84IEE309 pKa = 4.23PLALLLPMVLTTSLPIFVGQNWAANKK335 pKa = 9.41SLRR338 pKa = 11.84VRR340 pKa = 11.84RR341 pKa = 11.84GIKK344 pKa = 9.38QAIAACLVWQSVIALVLFWGADD366 pKa = 3.23MLGVGFCKK374 pKa = 10.35QSFVSQSIEE383 pKa = 3.56LAMCILPVSYY393 pKa = 10.54AALAGVMLYY402 pKa = 9.68VSCCNAIGRR411 pKa = 11.84SALALNVTLVRR422 pKa = 11.84LFVLSLPCAYY432 pKa = 10.52LGAQLNGFQGIVWALALANFVVGAWLLFRR461 pKa = 11.84ALNVQRR467 pKa = 11.84RR468 pKa = 11.84PQVLTMSS475 pKa = 4.17
MM1 pKa = 7.57GSDD4 pKa = 3.52NSPALSLGEE13 pKa = 3.94PLQVCDD19 pKa = 4.09KK20 pKa = 9.45QRR22 pKa = 11.84KK23 pKa = 7.32RR24 pKa = 11.84AVRR27 pKa = 11.84KK28 pKa = 9.07RR29 pKa = 11.84WILTGSTSAVLKK41 pKa = 10.71QLTLPMLWAILALFSADD58 pKa = 4.51LIEE61 pKa = 5.65LYY63 pKa = 10.22FASRR67 pKa = 11.84LGVEE71 pKa = 4.71EE72 pKa = 4.12LTAMSFTLPVQATLFAFAIGLGIVVATRR100 pKa = 11.84LTQAKK105 pKa = 9.27EE106 pKa = 4.14VEE108 pKa = 4.12QLAAVSLIFTVLVGATLAAVIWLGLQPLLTLLGFAEE144 pKa = 5.1FAARR148 pKa = 11.84EE149 pKa = 4.25QVWPTLEE156 pKa = 5.32RR157 pKa = 11.84YY158 pKa = 6.77MQYY161 pKa = 11.22RR162 pKa = 11.84LGAVVFFFIVMVVFGVLRR180 pKa = 11.84AFGNMRR186 pKa = 11.84GAAQLLVTFAGLQIVISAALFTPWAHH212 pKa = 6.27SALPLSGLEE221 pKa = 4.05RR222 pKa = 11.84LGVAHH227 pKa = 7.12FIAAVSASLYY237 pKa = 10.92ALYY240 pKa = 10.83LLRR243 pKa = 11.84IKK245 pKa = 10.68EE246 pKa = 4.19NISLKK251 pKa = 10.33VRR253 pKa = 11.84LLSQRR258 pKa = 11.84SRR260 pKa = 11.84QAFRR264 pKa = 11.84RR265 pKa = 11.84LFRR268 pKa = 11.84LFVPVVAMQLLTPLAQSLLMMIVASQGSDD297 pKa = 2.62AVAAFGVVMRR307 pKa = 11.84IEE309 pKa = 4.23PLALLLPMVLTTSLPIFVGQNWAANKK335 pKa = 9.41SLRR338 pKa = 11.84VRR340 pKa = 11.84RR341 pKa = 11.84GIKK344 pKa = 9.38QAIAACLVWQSVIALVLFWGADD366 pKa = 3.23MLGVGFCKK374 pKa = 10.35QSFVSQSIEE383 pKa = 3.56LAMCILPVSYY393 pKa = 10.54AALAGVMLYY402 pKa = 9.68VSCCNAIGRR411 pKa = 11.84SALALNVTLVRR422 pKa = 11.84LFVLSLPCAYY432 pKa = 10.52LGAQLNGFQGIVWALALANFVVGAWLLFRR461 pKa = 11.84ALNVQRR467 pKa = 11.84RR468 pKa = 11.84PQVLTMSS475 pKa = 4.17
Molecular weight: 51.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1545420 |
37 |
6585 |
352.0 |
39.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.37 ± 0.039 | 1.061 ± 0.012 |
5.632 ± 0.032 | 6.011 ± 0.032 |
4.07 ± 0.025 | 6.636 ± 0.035 |
2.432 ± 0.022 | 5.587 ± 0.029 |
4.804 ± 0.031 | 10.753 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.322 ± 0.019 | 4.064 ± 0.027 |
3.941 ± 0.025 | 5.385 ± 0.039 |
4.553 ± 0.031 | 6.633 ± 0.042 |
5.428 ± 0.037 | 6.861 ± 0.032 |
1.214 ± 0.015 | 3.243 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
