
Cyclovirus Equ1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Horse associated cyclovirus 1
Average proteome isoelectric point is 8.92
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
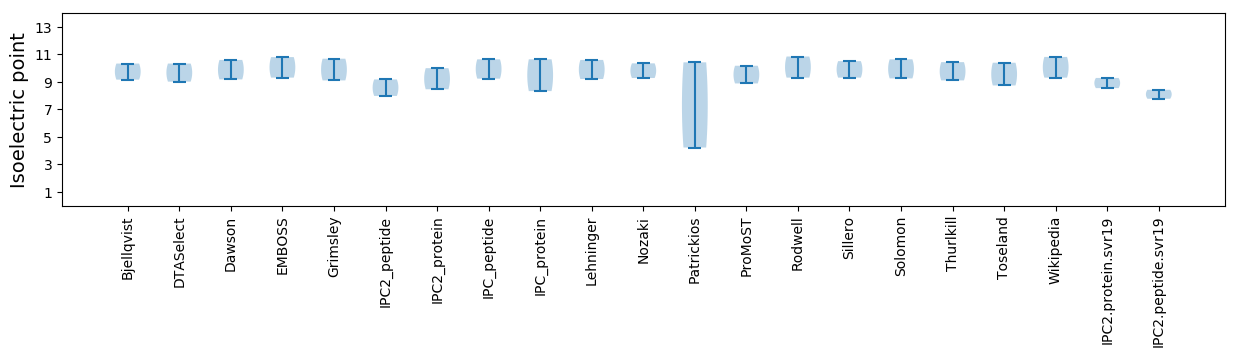
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4AQY4|A0A0H4AQY4_9CIRC Cap protein OS=Cyclovirus Equ1 OX=1673638 PE=4 SV=1
MM1 pKa = 7.14AQKK4 pKa = 10.5KK5 pKa = 8.79GWCFTYY11 pKa = 10.94NNYY14 pKa = 10.43DD15 pKa = 3.63EE16 pKa = 4.34QTLNGIFNRR25 pKa = 11.84CEE27 pKa = 4.44DD28 pKa = 3.15ICKK31 pKa = 10.24FIVVGKK37 pKa = 7.23EE38 pKa = 3.58VCPTTGTKK46 pKa = 9.95HH47 pKa = 5.17LQGYY51 pKa = 8.83CLFNEE56 pKa = 4.84RR57 pKa = 11.84KK58 pKa = 9.53RR59 pKa = 11.84FASVKK64 pKa = 9.34EE65 pKa = 4.03AIGGKK70 pKa = 7.46SHH72 pKa = 7.34IEE74 pKa = 3.76PAKK77 pKa = 10.59GSPQQNYY84 pKa = 10.28DD85 pKa = 3.36YY86 pKa = 9.17CTKK89 pKa = 10.79SGDD92 pKa = 3.52YY93 pKa = 7.22EE94 pKa = 4.42TRR96 pKa = 11.84GTLPKK101 pKa = 10.45GKK103 pKa = 10.39YY104 pKa = 6.88STLYY108 pKa = 8.96MVCEE112 pKa = 4.16EE113 pKa = 4.83LKK115 pKa = 10.91SGTPLRR121 pKa = 11.84TIALNEE127 pKa = 3.79PEE129 pKa = 4.46TFVRR133 pKa = 11.84HH134 pKa = 5.14HH135 pKa = 7.13RR136 pKa = 11.84GLQRR140 pKa = 11.84FTEE143 pKa = 4.12ILGEE147 pKa = 4.15STLRR151 pKa = 11.84DD152 pKa = 3.24WKK154 pKa = 9.99TEE156 pKa = 3.87VIVYY160 pKa = 10.04IGEE163 pKa = 4.26TGTGKK168 pKa = 10.0SRR170 pKa = 11.84KK171 pKa = 9.12CAEE174 pKa = 4.41LCGSTPTYY182 pKa = 9.94YY183 pKa = 10.58KK184 pKa = 10.51PRR186 pKa = 11.84GKK188 pKa = 8.93WWDD191 pKa = 3.72GYY193 pKa = 8.98TGQPNVIIDD202 pKa = 4.79DD203 pKa = 4.42FYY205 pKa = 11.73GWIKK209 pKa = 10.7YY210 pKa = 10.25DD211 pKa = 3.43EE212 pKa = 4.66LLRR215 pKa = 11.84ITDD218 pKa = 4.48RR219 pKa = 11.84YY220 pKa = 7.5PHH222 pKa = 6.44RR223 pKa = 11.84VPVKK227 pKa = 10.29FGYY230 pKa = 8.96QQFLAKK236 pKa = 9.85TIYY239 pKa = 8.79ITSNKK244 pKa = 8.51PMKK247 pKa = 9.66EE248 pKa = 3.46WYY250 pKa = 9.05TFDD253 pKa = 3.12ISALEE258 pKa = 3.76RR259 pKa = 11.84RR260 pKa = 11.84ITNVTLFKK268 pKa = 10.72KK269 pKa = 10.83LKK271 pKa = 9.92
MM1 pKa = 7.14AQKK4 pKa = 10.5KK5 pKa = 8.79GWCFTYY11 pKa = 10.94NNYY14 pKa = 10.43DD15 pKa = 3.63EE16 pKa = 4.34QTLNGIFNRR25 pKa = 11.84CEE27 pKa = 4.44DD28 pKa = 3.15ICKK31 pKa = 10.24FIVVGKK37 pKa = 7.23EE38 pKa = 3.58VCPTTGTKK46 pKa = 9.95HH47 pKa = 5.17LQGYY51 pKa = 8.83CLFNEE56 pKa = 4.84RR57 pKa = 11.84KK58 pKa = 9.53RR59 pKa = 11.84FASVKK64 pKa = 9.34EE65 pKa = 4.03AIGGKK70 pKa = 7.46SHH72 pKa = 7.34IEE74 pKa = 3.76PAKK77 pKa = 10.59GSPQQNYY84 pKa = 10.28DD85 pKa = 3.36YY86 pKa = 9.17CTKK89 pKa = 10.79SGDD92 pKa = 3.52YY93 pKa = 7.22EE94 pKa = 4.42TRR96 pKa = 11.84GTLPKK101 pKa = 10.45GKK103 pKa = 10.39YY104 pKa = 6.88STLYY108 pKa = 8.96MVCEE112 pKa = 4.16EE113 pKa = 4.83LKK115 pKa = 10.91SGTPLRR121 pKa = 11.84TIALNEE127 pKa = 3.79PEE129 pKa = 4.46TFVRR133 pKa = 11.84HH134 pKa = 5.14HH135 pKa = 7.13RR136 pKa = 11.84GLQRR140 pKa = 11.84FTEE143 pKa = 4.12ILGEE147 pKa = 4.15STLRR151 pKa = 11.84DD152 pKa = 3.24WKK154 pKa = 9.99TEE156 pKa = 3.87VIVYY160 pKa = 10.04IGEE163 pKa = 4.26TGTGKK168 pKa = 10.0SRR170 pKa = 11.84KK171 pKa = 9.12CAEE174 pKa = 4.41LCGSTPTYY182 pKa = 9.94YY183 pKa = 10.58KK184 pKa = 10.51PRR186 pKa = 11.84GKK188 pKa = 8.93WWDD191 pKa = 3.72GYY193 pKa = 8.98TGQPNVIIDD202 pKa = 4.79DD203 pKa = 4.42FYY205 pKa = 11.73GWIKK209 pKa = 10.7YY210 pKa = 10.25DD211 pKa = 3.43EE212 pKa = 4.66LLRR215 pKa = 11.84ITDD218 pKa = 4.48RR219 pKa = 11.84YY220 pKa = 7.5PHH222 pKa = 6.44RR223 pKa = 11.84VPVKK227 pKa = 10.29FGYY230 pKa = 8.96QQFLAKK236 pKa = 9.85TIYY239 pKa = 8.79ITSNKK244 pKa = 8.51PMKK247 pKa = 9.66EE248 pKa = 3.46WYY250 pKa = 9.05TFDD253 pKa = 3.12ISALEE258 pKa = 3.76RR259 pKa = 11.84RR260 pKa = 11.84ITNVTLFKK268 pKa = 10.72KK269 pKa = 10.83LKK271 pKa = 9.92
Molecular weight: 31.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4AQY4|A0A0H4AQY4_9CIRC Cap protein OS=Cyclovirus Equ1 OX=1673638 PE=4 SV=1
MM1 pKa = 7.63AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TYY9 pKa = 10.3RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84LPRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84YY19 pKa = 10.19VITKK23 pKa = 9.11RR24 pKa = 11.84FRR26 pKa = 11.84PRR28 pKa = 11.84YY29 pKa = 7.59QRR31 pKa = 11.84QRR33 pKa = 11.84YY34 pKa = 6.14RR35 pKa = 11.84TRR37 pKa = 11.84KK38 pKa = 8.69QLNAGDD44 pKa = 3.98KK45 pKa = 9.97YY46 pKa = 8.44FTRR49 pKa = 11.84MHH51 pKa = 6.48YY52 pKa = 10.37SSEE55 pKa = 3.7LRR57 pKa = 11.84IVANDD62 pKa = 3.36PSTYY66 pKa = 9.54SRR68 pKa = 11.84QMRR71 pKa = 11.84IYY73 pKa = 9.62MSWFSNFIDD82 pKa = 5.5FNQNWSQYY90 pKa = 8.22KK91 pKa = 6.82VHH93 pKa = 6.83KK94 pKa = 9.94IVMKK98 pKa = 7.2FTPMFNNTQVNDD110 pKa = 3.52AVGNYY115 pKa = 9.03AVAVMNTGLGFGGTDD130 pKa = 3.44EE131 pKa = 4.39NSSTIIEE138 pKa = 4.47EE139 pKa = 3.8NRR141 pKa = 11.84MSYY144 pKa = 9.43EE145 pKa = 3.7QLVAIPSTKK154 pKa = 9.22IRR156 pKa = 11.84KK157 pKa = 6.38ATKK160 pKa = 10.06AVTLVFKK167 pKa = 10.4PKK169 pKa = 9.82IANTTGVVLGEE180 pKa = 3.9SSYY183 pKa = 10.2VHH185 pKa = 6.35NNNLIYY191 pKa = 10.48HH192 pKa = 6.48PWISTLRR199 pKa = 11.84ASTNANKK206 pKa = 9.6VPQLYY211 pKa = 9.75GAYY214 pKa = 9.52FAHH217 pKa = 6.74QGTANEE223 pKa = 4.49AVPQEE228 pKa = 4.12INYY231 pKa = 8.64MVDD234 pKa = 2.51VFAYY238 pKa = 8.67VTFKK242 pKa = 10.87GYY244 pKa = 10.56RR245 pKa = 11.84LPVSSKK251 pKa = 10.29QKK253 pKa = 9.79IRR255 pKa = 11.84QLNPVPEE262 pKa = 4.94PDD264 pKa = 3.41MTDD267 pKa = 2.82MM268 pKa = 5.7
MM1 pKa = 7.63AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TYY9 pKa = 10.3RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84LPRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84YY19 pKa = 10.19VITKK23 pKa = 9.11RR24 pKa = 11.84FRR26 pKa = 11.84PRR28 pKa = 11.84YY29 pKa = 7.59QRR31 pKa = 11.84QRR33 pKa = 11.84YY34 pKa = 6.14RR35 pKa = 11.84TRR37 pKa = 11.84KK38 pKa = 8.69QLNAGDD44 pKa = 3.98KK45 pKa = 9.97YY46 pKa = 8.44FTRR49 pKa = 11.84MHH51 pKa = 6.48YY52 pKa = 10.37SSEE55 pKa = 3.7LRR57 pKa = 11.84IVANDD62 pKa = 3.36PSTYY66 pKa = 9.54SRR68 pKa = 11.84QMRR71 pKa = 11.84IYY73 pKa = 9.62MSWFSNFIDD82 pKa = 5.5FNQNWSQYY90 pKa = 8.22KK91 pKa = 6.82VHH93 pKa = 6.83KK94 pKa = 9.94IVMKK98 pKa = 7.2FTPMFNNTQVNDD110 pKa = 3.52AVGNYY115 pKa = 9.03AVAVMNTGLGFGGTDD130 pKa = 3.44EE131 pKa = 4.39NSSTIIEE138 pKa = 4.47EE139 pKa = 3.8NRR141 pKa = 11.84MSYY144 pKa = 9.43EE145 pKa = 3.7QLVAIPSTKK154 pKa = 9.22IRR156 pKa = 11.84KK157 pKa = 6.38ATKK160 pKa = 10.06AVTLVFKK167 pKa = 10.4PKK169 pKa = 9.82IANTTGVVLGEE180 pKa = 3.9SSYY183 pKa = 10.2VHH185 pKa = 6.35NNNLIYY191 pKa = 10.48HH192 pKa = 6.48PWISTLRR199 pKa = 11.84ASTNANKK206 pKa = 9.6VPQLYY211 pKa = 9.75GAYY214 pKa = 9.52FAHH217 pKa = 6.74QGTANEE223 pKa = 4.49AVPQEE228 pKa = 4.12INYY231 pKa = 8.64MVDD234 pKa = 2.51VFAYY238 pKa = 8.67VTFKK242 pKa = 10.87GYY244 pKa = 10.56RR245 pKa = 11.84LPVSSKK251 pKa = 10.29QKK253 pKa = 9.79IRR255 pKa = 11.84QLNPVPEE262 pKa = 4.94PDD264 pKa = 3.41MTDD267 pKa = 2.82MM268 pKa = 5.7
Molecular weight: 31.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
539 |
268 |
271 |
269.5 |
31.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.638 ± 1.215 | 1.67 ± 1.19 |
3.525 ± 0.385 | 5.195 ± 1.308 |
4.638 ± 0.151 | 6.308 ± 1.57 |
1.855 ± 0.007 | 5.751 ± 0.376 |
7.607 ± 1.432 | 5.566 ± 0.775 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 1.074 | 5.751 ± 1.485 |
4.824 ± 0.285 | 4.082 ± 0.548 |
7.792 ± 1.36 | 5.195 ± 0.818 |
8.534 ± 0.763 | 6.122 ± 1.221 |
1.67 ± 0.392 | 6.679 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
