
Macaca fascicularis papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 6
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
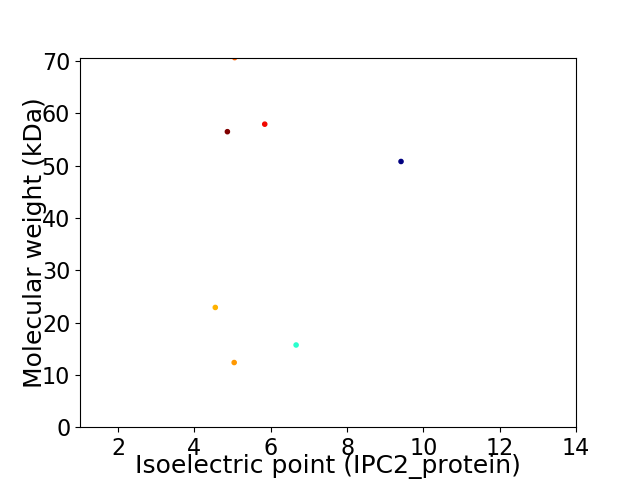
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
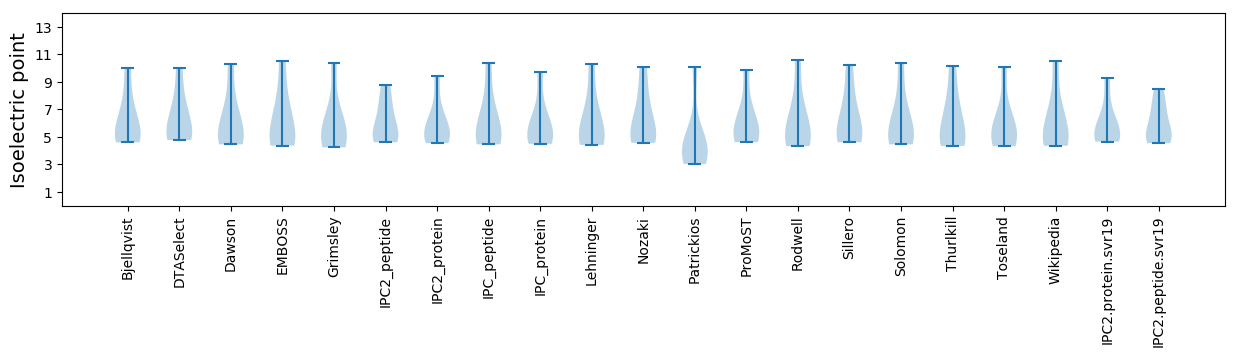
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8QPP9|F8QPP9_9PAPI Minor capsid protein L2 OS=Macaca fascicularis papillomavirus 2 OX=915424 GN=L2 PE=3 SV=1
MM1 pKa = 7.32LHH3 pKa = 6.87LGLMQRR9 pKa = 11.84DD10 pKa = 3.91MVKK13 pKa = 10.64LDD15 pKa = 3.66FGRR18 pKa = 11.84LSIIRR23 pKa = 11.84TLSLLLSPVQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84DD37 pKa = 3.62RR38 pKa = 11.84DD39 pKa = 3.69HH40 pKa = 6.94QPPPTPTPQKK50 pKa = 10.94DD51 pKa = 3.31PNTPLKK57 pKa = 9.19WYY59 pKa = 10.3QSIRR63 pKa = 11.84PPPLPPQPPLPPLNSEE79 pKa = 4.21PEE81 pKa = 4.33GTGEE85 pKa = 3.93KK86 pKa = 10.43PPVQEE91 pKa = 4.98EE92 pKa = 4.38EE93 pKa = 4.48NEE95 pKa = 3.93DD96 pKa = 3.48TRR98 pKa = 11.84PSKK101 pKa = 9.47KK102 pKa = 8.73TKK104 pKa = 10.28EE105 pKa = 4.34NGDD108 pKa = 3.55HH109 pKa = 6.43TSGDD113 pKa = 3.9GEE115 pKa = 4.54KK116 pKa = 10.64EE117 pKa = 4.18GGDD120 pKa = 3.95PDD122 pKa = 4.94PGPVPHH128 pKa = 7.82PDD130 pKa = 4.02PDD132 pKa = 4.71PDD134 pKa = 4.76PDD136 pKa = 4.38PEE138 pKa = 4.78PEE140 pKa = 4.65PEE142 pKa = 4.87PDD144 pKa = 4.7PNPQPQPIPAPDD156 pKa = 4.7PDD158 pKa = 4.69LDD160 pKa = 4.46PGQVASFLEE169 pKa = 5.05GVASYY174 pKa = 9.04LHH176 pKa = 6.4RR177 pKa = 11.84WEE179 pKa = 4.56VGYY182 pKa = 9.67QQLVKK187 pKa = 10.47DD188 pKa = 4.08IKK190 pKa = 10.91EE191 pKa = 4.24DD192 pKa = 4.16LDD194 pKa = 4.21NYY196 pKa = 8.24WLKK199 pKa = 11.0LQTPQQ204 pKa = 4.22
MM1 pKa = 7.32LHH3 pKa = 6.87LGLMQRR9 pKa = 11.84DD10 pKa = 3.91MVKK13 pKa = 10.64LDD15 pKa = 3.66FGRR18 pKa = 11.84LSIIRR23 pKa = 11.84TLSLLLSPVQRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84DD37 pKa = 3.62RR38 pKa = 11.84DD39 pKa = 3.69HH40 pKa = 6.94QPPPTPTPQKK50 pKa = 10.94DD51 pKa = 3.31PNTPLKK57 pKa = 9.19WYY59 pKa = 10.3QSIRR63 pKa = 11.84PPPLPPQPPLPPLNSEE79 pKa = 4.21PEE81 pKa = 4.33GTGEE85 pKa = 3.93KK86 pKa = 10.43PPVQEE91 pKa = 4.98EE92 pKa = 4.38EE93 pKa = 4.48NEE95 pKa = 3.93DD96 pKa = 3.48TRR98 pKa = 11.84PSKK101 pKa = 9.47KK102 pKa = 8.73TKK104 pKa = 10.28EE105 pKa = 4.34NGDD108 pKa = 3.55HH109 pKa = 6.43TSGDD113 pKa = 3.9GEE115 pKa = 4.54KK116 pKa = 10.64EE117 pKa = 4.18GGDD120 pKa = 3.95PDD122 pKa = 4.94PGPVPHH128 pKa = 7.82PDD130 pKa = 4.02PDD132 pKa = 4.71PDD134 pKa = 4.76PDD136 pKa = 4.38PEE138 pKa = 4.78PEE140 pKa = 4.65PEE142 pKa = 4.87PDD144 pKa = 4.7PNPQPQPIPAPDD156 pKa = 4.7PDD158 pKa = 4.69LDD160 pKa = 4.46PGQVASFLEE169 pKa = 5.05GVASYY174 pKa = 9.04LHH176 pKa = 6.4RR177 pKa = 11.84WEE179 pKa = 4.56VGYY182 pKa = 9.67QQLVKK187 pKa = 10.47DD188 pKa = 4.08IKK190 pKa = 10.91EE191 pKa = 4.24DD192 pKa = 4.16LDD194 pKa = 4.21NYY196 pKa = 8.24WLKK199 pKa = 11.0LQTPQQ204 pKa = 4.22
Molecular weight: 22.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8QPP8|F8QPP8_9PAPI E4 OS=Macaca fascicularis papillomavirus 2 OX=915424 GN=E4 PE=4 SV=1
MM1 pKa = 7.02EE2 pKa = 4.44TLSEE6 pKa = 4.19RR7 pKa = 11.84FHH9 pKa = 7.09ALQEE13 pKa = 4.13KK14 pKa = 10.35LMEE17 pKa = 4.88IYY19 pKa = 10.51EE20 pKa = 4.34SGKK23 pKa = 8.49NTIAAQIEE31 pKa = 4.3HH32 pKa = 6.22WQLLRR37 pKa = 11.84QEE39 pKa = 4.16QVLLHH44 pKa = 6.15FARR47 pKa = 11.84KK48 pKa = 9.64RR49 pKa = 11.84GVTRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPNLAITEE66 pKa = 4.17QKK68 pKa = 10.94AKK70 pKa = 10.68DD71 pKa = 3.99AIGMGLLLQSLEE83 pKa = 3.82KK84 pKa = 10.53SPYY87 pKa = 10.15GGEE90 pKa = 3.9SWSLVDD96 pKa = 4.03TSLEE100 pKa = 4.27TFRR103 pKa = 11.84SPPQQCFKK111 pKa = 11.06KK112 pKa = 10.31GGQEE116 pKa = 3.79VEE118 pKa = 4.09VRR120 pKa = 11.84FDD122 pKa = 3.81GDD124 pKa = 3.54PEE126 pKa = 5.48NIMLYY131 pKa = 9.73TSWQHH136 pKa = 7.26IYY138 pKa = 10.37FQDD141 pKa = 4.85SHH143 pKa = 8.03DD144 pKa = 3.76SWQKK148 pKa = 9.55VQGQVDD154 pKa = 4.2YY155 pKa = 11.62DD156 pKa = 3.56GLFYY160 pKa = 10.79FDD162 pKa = 4.98GPVKK166 pKa = 10.49VYY168 pKa = 10.68YY169 pKa = 9.34VTFGTDD175 pKa = 2.57AKK177 pKa = 10.61RR178 pKa = 11.84YY179 pKa = 8.62GKK181 pKa = 8.54TGLWEE186 pKa = 3.8VVYY189 pKa = 11.08NKK191 pKa = 10.4DD192 pKa = 3.68IIFAPVTSSTPEE204 pKa = 4.17GQGSPAPADD213 pKa = 3.62SNTAEE218 pKa = 4.78GSQHH222 pKa = 4.79PTEE225 pKa = 4.42VVSVDD230 pKa = 3.3SATTSAPAATSATAKK245 pKa = 9.77QRR247 pKa = 11.84ARR249 pKa = 11.84RR250 pKa = 11.84YY251 pKa = 8.85GRR253 pKa = 11.84KK254 pKa = 9.39ASSPGGGEE262 pKa = 3.61RR263 pKa = 11.84GYY265 pKa = 11.34SPLKK269 pKa = 8.94EE270 pKa = 4.21NQRR273 pKa = 11.84EE274 pKa = 4.11RR275 pKa = 11.84RR276 pKa = 11.84PHH278 pKa = 4.85KK279 pKa = 9.9RR280 pKa = 11.84GRR282 pKa = 11.84GEE284 pKa = 3.5RR285 pKa = 11.84GGRR288 pKa = 11.84SRR290 pKa = 11.84SRR292 pKa = 11.84SRR294 pKa = 11.84ATSRR298 pKa = 11.84SRR300 pKa = 11.84SRR302 pKa = 11.84SRR304 pKa = 11.84SRR306 pKa = 11.84TRR308 pKa = 11.84TRR310 pKa = 11.84ARR312 pKa = 11.84PQSATTTHH320 pKa = 6.9PCTRR324 pKa = 11.84SRR326 pKa = 11.84SRR328 pKa = 11.84SRR330 pKa = 11.84PGCVLSGGGGVLPSQVGSRR349 pKa = 11.84LSTVGQGHH357 pKa = 5.81QGRR360 pKa = 11.84LGQLLAEE367 pKa = 4.56ATDD370 pKa = 3.88PPVILLRR377 pKa = 11.84GDD379 pKa = 3.61PNAVKK384 pKa = 10.59CFRR387 pKa = 11.84FRR389 pKa = 11.84AKK391 pKa = 10.45DD392 pKa = 3.36KK393 pKa = 11.24YY394 pKa = 9.79RR395 pKa = 11.84GLYY398 pKa = 9.2TRR400 pKa = 11.84MSTTWSWVGADD411 pKa = 3.24NNDD414 pKa = 3.91RR415 pKa = 11.84IGRR418 pKa = 11.84ARR420 pKa = 11.84MLVAFSSYY428 pKa = 6.95EE429 pKa = 3.81QRR431 pKa = 11.84HH432 pKa = 5.89LFLKK436 pKa = 9.87HH437 pKa = 6.01LKK439 pKa = 9.61VPKK442 pKa = 10.66GMDD445 pKa = 2.83WSLGQFDD452 pKa = 4.27SLL454 pKa = 4.55
MM1 pKa = 7.02EE2 pKa = 4.44TLSEE6 pKa = 4.19RR7 pKa = 11.84FHH9 pKa = 7.09ALQEE13 pKa = 4.13KK14 pKa = 10.35LMEE17 pKa = 4.88IYY19 pKa = 10.51EE20 pKa = 4.34SGKK23 pKa = 8.49NTIAAQIEE31 pKa = 4.3HH32 pKa = 6.22WQLLRR37 pKa = 11.84QEE39 pKa = 4.16QVLLHH44 pKa = 6.15FARR47 pKa = 11.84KK48 pKa = 9.64RR49 pKa = 11.84GVTRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPNLAITEE66 pKa = 4.17QKK68 pKa = 10.94AKK70 pKa = 10.68DD71 pKa = 3.99AIGMGLLLQSLEE83 pKa = 3.82KK84 pKa = 10.53SPYY87 pKa = 10.15GGEE90 pKa = 3.9SWSLVDD96 pKa = 4.03TSLEE100 pKa = 4.27TFRR103 pKa = 11.84SPPQQCFKK111 pKa = 11.06KK112 pKa = 10.31GGQEE116 pKa = 3.79VEE118 pKa = 4.09VRR120 pKa = 11.84FDD122 pKa = 3.81GDD124 pKa = 3.54PEE126 pKa = 5.48NIMLYY131 pKa = 9.73TSWQHH136 pKa = 7.26IYY138 pKa = 10.37FQDD141 pKa = 4.85SHH143 pKa = 8.03DD144 pKa = 3.76SWQKK148 pKa = 9.55VQGQVDD154 pKa = 4.2YY155 pKa = 11.62DD156 pKa = 3.56GLFYY160 pKa = 10.79FDD162 pKa = 4.98GPVKK166 pKa = 10.49VYY168 pKa = 10.68YY169 pKa = 9.34VTFGTDD175 pKa = 2.57AKK177 pKa = 10.61RR178 pKa = 11.84YY179 pKa = 8.62GKK181 pKa = 8.54TGLWEE186 pKa = 3.8VVYY189 pKa = 11.08NKK191 pKa = 10.4DD192 pKa = 3.68IIFAPVTSSTPEE204 pKa = 4.17GQGSPAPADD213 pKa = 3.62SNTAEE218 pKa = 4.78GSQHH222 pKa = 4.79PTEE225 pKa = 4.42VVSVDD230 pKa = 3.3SATTSAPAATSATAKK245 pKa = 9.77QRR247 pKa = 11.84ARR249 pKa = 11.84RR250 pKa = 11.84YY251 pKa = 8.85GRR253 pKa = 11.84KK254 pKa = 9.39ASSPGGGEE262 pKa = 3.61RR263 pKa = 11.84GYY265 pKa = 11.34SPLKK269 pKa = 8.94EE270 pKa = 4.21NQRR273 pKa = 11.84EE274 pKa = 4.11RR275 pKa = 11.84RR276 pKa = 11.84PHH278 pKa = 4.85KK279 pKa = 9.9RR280 pKa = 11.84GRR282 pKa = 11.84GEE284 pKa = 3.5RR285 pKa = 11.84GGRR288 pKa = 11.84SRR290 pKa = 11.84SRR292 pKa = 11.84SRR294 pKa = 11.84ATSRR298 pKa = 11.84SRR300 pKa = 11.84SRR302 pKa = 11.84SRR304 pKa = 11.84SRR306 pKa = 11.84TRR308 pKa = 11.84TRR310 pKa = 11.84ARR312 pKa = 11.84PQSATTTHH320 pKa = 6.9PCTRR324 pKa = 11.84SRR326 pKa = 11.84SRR328 pKa = 11.84SRR330 pKa = 11.84PGCVLSGGGGVLPSQVGSRR349 pKa = 11.84LSTVGQGHH357 pKa = 5.81QGRR360 pKa = 11.84LGQLLAEE367 pKa = 4.56ATDD370 pKa = 3.88PPVILLRR377 pKa = 11.84GDD379 pKa = 3.61PNAVKK384 pKa = 10.59CFRR387 pKa = 11.84FRR389 pKa = 11.84AKK391 pKa = 10.45DD392 pKa = 3.36KK393 pKa = 11.24YY394 pKa = 9.79RR395 pKa = 11.84GLYY398 pKa = 9.2TRR400 pKa = 11.84MSTTWSWVGADD411 pKa = 3.24NNDD414 pKa = 3.91RR415 pKa = 11.84IGRR418 pKa = 11.84ARR420 pKa = 11.84MLVAFSSYY428 pKa = 6.95EE429 pKa = 3.81QRR431 pKa = 11.84HH432 pKa = 5.89LFLKK436 pKa = 9.87HH437 pKa = 6.01LKK439 pKa = 9.61VPKK442 pKa = 10.66GMDD445 pKa = 2.83WSLGQFDD452 pKa = 4.27SLL454 pKa = 4.55
Molecular weight: 50.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2558 |
108 |
614 |
365.4 |
40.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.059 ± 0.691 | 1.798 ± 0.587 |
6.568 ± 0.672 | 6.177 ± 0.953 |
4.027 ± 0.43 | 7.232 ± 0.759 |
1.916 ± 0.226 | 4.183 ± 0.419 |
3.987 ± 0.568 | 8.913 ± 0.512 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.446 ± 0.297 | 3.675 ± 0.792 |
6.88 ± 1.344 | 5.238 ± 0.274 |
7.271 ± 0.645 | 7.545 ± 0.799 |
6.138 ± 0.577 | 6.333 ± 0.554 |
1.407 ± 0.35 | 3.206 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
