
Adlercreutzia caecicola
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Parvibacter
Average proteome isoelectric point is 5.73
Get precalculated fractions of proteins
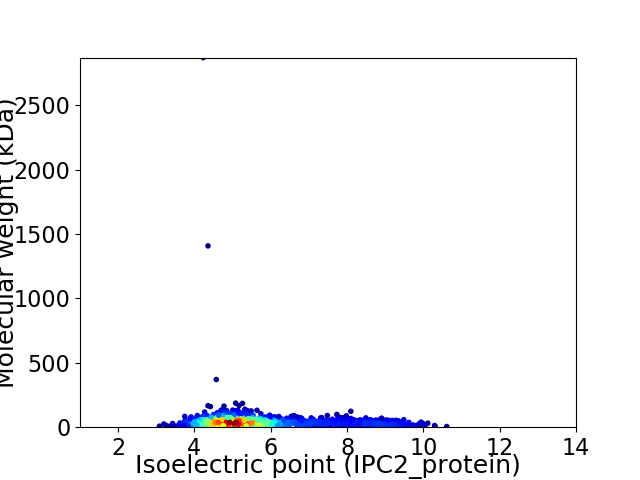
Virtual 2D-PAGE plot for 1821 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4T9TEF9|A0A4T9TEF9_9ACTN VanZ domain-containing protein OS=Adlercreutzia caecicola OX=747645 GN=E5982_02910 PE=4 SV=1
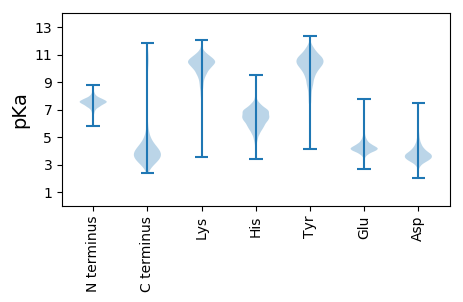
MM1 pKa = 7.15TRR3 pKa = 11.84QDD5 pKa = 3.77EE6 pKa = 4.17FDD8 pKa = 3.2AAARR12 pKa = 11.84ALRR15 pKa = 11.84ADD17 pKa = 4.33AEE19 pKa = 4.3QHH21 pKa = 5.97PVPASLQPEE30 pKa = 4.62AIEE33 pKa = 4.77AMLLAHH39 pKa = 7.47DD40 pKa = 4.42AQKK43 pKa = 11.23SDD45 pKa = 4.2GEE47 pKa = 4.58TADD50 pKa = 3.42SGQPASAAPAPVEE63 pKa = 4.27TGSAAAGTASPRR75 pKa = 11.84RR76 pKa = 11.84HH77 pKa = 5.01VTWRR81 pKa = 11.84WPIAAAACLALLCGVGVVGALAAQGKK107 pKa = 8.62FSPAAIDD114 pKa = 4.16APSSPLPAVSDD125 pKa = 4.22PSAPASPEE133 pKa = 4.14SGLSTAQGYY142 pKa = 8.72AQVRR146 pKa = 11.84DD147 pKa = 4.08CLLASQQAMGDD158 pKa = 3.43GGTMFMEE165 pKa = 5.3DD166 pKa = 2.95KK167 pKa = 9.88TAGVARR173 pKa = 11.84TEE175 pKa = 3.97TATAFTAQNSTKK187 pKa = 10.43DD188 pKa = 3.46SSFTDD193 pKa = 3.3TNEE196 pKa = 3.71RR197 pKa = 11.84TEE199 pKa = 4.81GIGEE203 pKa = 4.28ADD205 pKa = 3.53VAKK208 pKa = 10.16TDD210 pKa = 3.99GRR212 pKa = 11.84WLYY215 pKa = 11.12ALQDD219 pKa = 3.28SGTTVAIVDD228 pKa = 3.84PAEE231 pKa = 4.05GHH233 pKa = 5.17MTPAGAIEE241 pKa = 4.31APKK244 pKa = 9.8GAQVSEE250 pKa = 5.06FYY252 pKa = 11.33VEE254 pKa = 4.82DD255 pKa = 3.45GRR257 pKa = 11.84LYY259 pKa = 11.04LLSTVFPEE267 pKa = 5.44GVEE270 pKa = 4.28QPDD273 pKa = 3.59GTYY276 pKa = 9.82TFPPEE281 pKa = 3.86TTRR284 pKa = 11.84LDD286 pKa = 3.87VYY288 pKa = 11.33DD289 pKa = 3.75VTDD292 pKa = 3.91PANPQATGLLTQTGTYY308 pKa = 7.85NTVRR312 pKa = 11.84FADD315 pKa = 4.54GYY317 pKa = 11.24VYY319 pKa = 10.8LFSDD323 pKa = 2.84QWTYY327 pKa = 10.57DD328 pKa = 3.36TSEE331 pKa = 4.1GTDD334 pKa = 3.25PSAYY338 pKa = 9.62VPCVNGAAVEE348 pKa = 4.31CGDD351 pKa = 4.37IYY353 pKa = 11.08LPPEE357 pKa = 4.15NMGDD361 pKa = 3.39HH362 pKa = 6.27YY363 pKa = 11.65LVITTVDD370 pKa = 3.22AANPSAVVDD379 pKa = 4.28QKK381 pKa = 11.7AILCNGGSVYY391 pKa = 10.12MSQDD395 pKa = 3.36SIFLYY400 pKa = 8.58EE401 pKa = 3.93TTGYY405 pKa = 10.01NWGIMPIARR414 pKa = 11.84FAADD418 pKa = 5.26DD419 pKa = 3.64MAEE422 pKa = 4.31PAGEE426 pKa = 3.98TDD428 pKa = 5.17PIAEE432 pKa = 4.84AATEE436 pKa = 4.23DD437 pKa = 4.46PGDD440 pKa = 3.94APADD444 pKa = 3.43EE445 pKa = 4.82SEE447 pKa = 4.48NGEE450 pKa = 4.23EE451 pKa = 4.44APGATSPAEE460 pKa = 4.34GPDD463 pKa = 3.81GEE465 pKa = 5.13GVNDD469 pKa = 4.71DD470 pKa = 4.52PAVDD474 pKa = 5.09DD475 pKa = 4.76PATLPADD482 pKa = 4.36DD483 pKa = 5.26PATLPADD490 pKa = 4.19GPAAEE495 pKa = 4.49PQSPEE500 pKa = 4.12TPDD503 pKa = 3.77EE504 pKa = 4.12EE505 pKa = 4.45AQAPSDD511 pKa = 3.91NSADD515 pKa = 3.74DD516 pKa = 4.08PSEE519 pKa = 4.98GEE521 pKa = 4.11ADD523 pKa = 3.85TPDD526 pKa = 3.45SSGGDD531 pKa = 3.57DD532 pKa = 4.85PDD534 pKa = 4.23LKK536 pKa = 10.02TCIRR540 pKa = 11.84KK541 pKa = 8.06FTFDD545 pKa = 3.21KK546 pKa = 11.24GQLEE550 pKa = 4.47GVAQGEE556 pKa = 4.78VNGTLDD562 pKa = 4.42SSFCLDD568 pKa = 3.79EE569 pKa = 4.25YY570 pKa = 11.28NGYY573 pKa = 10.02LRR575 pKa = 11.84VVTTVYY581 pKa = 11.21GEE583 pKa = 4.27DD584 pKa = 3.34TSSALTVLNEE594 pKa = 3.78QMEE597 pKa = 4.72EE598 pKa = 3.9VGNIADD604 pKa = 3.86VAPGEE609 pKa = 4.26QVYY612 pKa = 9.88SARR615 pKa = 11.84FMGNVGYY622 pKa = 9.93FVTFKK627 pKa = 10.84QVDD630 pKa = 3.6PLFSVDD636 pKa = 4.92LSNPEE641 pKa = 3.77NPTIIGQLKK650 pKa = 9.15IPGFSEE656 pKa = 4.05YY657 pKa = 10.73LHH659 pKa = 7.02PYY661 pKa = 9.62GEE663 pKa = 4.14NLLLGIGMSADD674 pKa = 3.52DD675 pKa = 5.02AGATDD680 pKa = 4.4GVKK683 pKa = 10.78LSMFDD688 pKa = 3.28TSNPADD694 pKa = 3.54VRR696 pKa = 11.84EE697 pKa = 4.21VATAVLDD704 pKa = 3.48DD705 pKa = 4.4TYY707 pKa = 11.68YY708 pKa = 11.18SDD710 pKa = 3.95VFNEE714 pKa = 3.58YY715 pKa = 9.81RR716 pKa = 11.84AALIDD721 pKa = 4.08PASDD725 pKa = 3.4MIGFPVSTDD734 pKa = 2.98RR735 pKa = 11.84EE736 pKa = 4.25AYY738 pKa = 9.4AVYY741 pKa = 9.88GYY743 pKa = 10.67SEE745 pKa = 5.35DD746 pKa = 3.9NGFSQHH752 pKa = 5.82MLEE755 pKa = 4.41EE756 pKa = 4.57TNGTGWVPTRR766 pKa = 11.84GIRR769 pKa = 11.84IGDD772 pKa = 3.33VLYY775 pKa = 10.79VVKK778 pKa = 10.85GNAAEE783 pKa = 4.25SYY785 pKa = 10.61RR786 pKa = 11.84IGDD789 pKa = 3.59YY790 pKa = 10.99QKK792 pKa = 10.6IDD794 pKa = 3.63DD795 pKa = 4.63LLII798 pKa = 4.67
MM1 pKa = 7.15TRR3 pKa = 11.84QDD5 pKa = 3.77EE6 pKa = 4.17FDD8 pKa = 3.2AAARR12 pKa = 11.84ALRR15 pKa = 11.84ADD17 pKa = 4.33AEE19 pKa = 4.3QHH21 pKa = 5.97PVPASLQPEE30 pKa = 4.62AIEE33 pKa = 4.77AMLLAHH39 pKa = 7.47DD40 pKa = 4.42AQKK43 pKa = 11.23SDD45 pKa = 4.2GEE47 pKa = 4.58TADD50 pKa = 3.42SGQPASAAPAPVEE63 pKa = 4.27TGSAAAGTASPRR75 pKa = 11.84RR76 pKa = 11.84HH77 pKa = 5.01VTWRR81 pKa = 11.84WPIAAAACLALLCGVGVVGALAAQGKK107 pKa = 8.62FSPAAIDD114 pKa = 4.16APSSPLPAVSDD125 pKa = 4.22PSAPASPEE133 pKa = 4.14SGLSTAQGYY142 pKa = 8.72AQVRR146 pKa = 11.84DD147 pKa = 4.08CLLASQQAMGDD158 pKa = 3.43GGTMFMEE165 pKa = 5.3DD166 pKa = 2.95KK167 pKa = 9.88TAGVARR173 pKa = 11.84TEE175 pKa = 3.97TATAFTAQNSTKK187 pKa = 10.43DD188 pKa = 3.46SSFTDD193 pKa = 3.3TNEE196 pKa = 3.71RR197 pKa = 11.84TEE199 pKa = 4.81GIGEE203 pKa = 4.28ADD205 pKa = 3.53VAKK208 pKa = 10.16TDD210 pKa = 3.99GRR212 pKa = 11.84WLYY215 pKa = 11.12ALQDD219 pKa = 3.28SGTTVAIVDD228 pKa = 3.84PAEE231 pKa = 4.05GHH233 pKa = 5.17MTPAGAIEE241 pKa = 4.31APKK244 pKa = 9.8GAQVSEE250 pKa = 5.06FYY252 pKa = 11.33VEE254 pKa = 4.82DD255 pKa = 3.45GRR257 pKa = 11.84LYY259 pKa = 11.04LLSTVFPEE267 pKa = 5.44GVEE270 pKa = 4.28QPDD273 pKa = 3.59GTYY276 pKa = 9.82TFPPEE281 pKa = 3.86TTRR284 pKa = 11.84LDD286 pKa = 3.87VYY288 pKa = 11.33DD289 pKa = 3.75VTDD292 pKa = 3.91PANPQATGLLTQTGTYY308 pKa = 7.85NTVRR312 pKa = 11.84FADD315 pKa = 4.54GYY317 pKa = 11.24VYY319 pKa = 10.8LFSDD323 pKa = 2.84QWTYY327 pKa = 10.57DD328 pKa = 3.36TSEE331 pKa = 4.1GTDD334 pKa = 3.25PSAYY338 pKa = 9.62VPCVNGAAVEE348 pKa = 4.31CGDD351 pKa = 4.37IYY353 pKa = 11.08LPPEE357 pKa = 4.15NMGDD361 pKa = 3.39HH362 pKa = 6.27YY363 pKa = 11.65LVITTVDD370 pKa = 3.22AANPSAVVDD379 pKa = 4.28QKK381 pKa = 11.7AILCNGGSVYY391 pKa = 10.12MSQDD395 pKa = 3.36SIFLYY400 pKa = 8.58EE401 pKa = 3.93TTGYY405 pKa = 10.01NWGIMPIARR414 pKa = 11.84FAADD418 pKa = 5.26DD419 pKa = 3.64MAEE422 pKa = 4.31PAGEE426 pKa = 3.98TDD428 pKa = 5.17PIAEE432 pKa = 4.84AATEE436 pKa = 4.23DD437 pKa = 4.46PGDD440 pKa = 3.94APADD444 pKa = 3.43EE445 pKa = 4.82SEE447 pKa = 4.48NGEE450 pKa = 4.23EE451 pKa = 4.44APGATSPAEE460 pKa = 4.34GPDD463 pKa = 3.81GEE465 pKa = 5.13GVNDD469 pKa = 4.71DD470 pKa = 4.52PAVDD474 pKa = 5.09DD475 pKa = 4.76PATLPADD482 pKa = 4.36DD483 pKa = 5.26PATLPADD490 pKa = 4.19GPAAEE495 pKa = 4.49PQSPEE500 pKa = 4.12TPDD503 pKa = 3.77EE504 pKa = 4.12EE505 pKa = 4.45AQAPSDD511 pKa = 3.91NSADD515 pKa = 3.74DD516 pKa = 4.08PSEE519 pKa = 4.98GEE521 pKa = 4.11ADD523 pKa = 3.85TPDD526 pKa = 3.45SSGGDD531 pKa = 3.57DD532 pKa = 4.85PDD534 pKa = 4.23LKK536 pKa = 10.02TCIRR540 pKa = 11.84KK541 pKa = 8.06FTFDD545 pKa = 3.21KK546 pKa = 11.24GQLEE550 pKa = 4.47GVAQGEE556 pKa = 4.78VNGTLDD562 pKa = 4.42SSFCLDD568 pKa = 3.79EE569 pKa = 4.25YY570 pKa = 11.28NGYY573 pKa = 10.02LRR575 pKa = 11.84VVTTVYY581 pKa = 11.21GEE583 pKa = 4.27DD584 pKa = 3.34TSSALTVLNEE594 pKa = 3.78QMEE597 pKa = 4.72EE598 pKa = 3.9VGNIADD604 pKa = 3.86VAPGEE609 pKa = 4.26QVYY612 pKa = 9.88SARR615 pKa = 11.84FMGNVGYY622 pKa = 9.93FVTFKK627 pKa = 10.84QVDD630 pKa = 3.6PLFSVDD636 pKa = 4.92LSNPEE641 pKa = 3.77NPTIIGQLKK650 pKa = 9.15IPGFSEE656 pKa = 4.05YY657 pKa = 10.73LHH659 pKa = 7.02PYY661 pKa = 9.62GEE663 pKa = 4.14NLLLGIGMSADD674 pKa = 3.52DD675 pKa = 5.02AGATDD680 pKa = 4.4GVKK683 pKa = 10.78LSMFDD688 pKa = 3.28TSNPADD694 pKa = 3.54VRR696 pKa = 11.84EE697 pKa = 4.21VATAVLDD704 pKa = 3.48DD705 pKa = 4.4TYY707 pKa = 11.68YY708 pKa = 11.18SDD710 pKa = 3.95VFNEE714 pKa = 3.58YY715 pKa = 9.81RR716 pKa = 11.84AALIDD721 pKa = 4.08PASDD725 pKa = 3.4MIGFPVSTDD734 pKa = 2.98RR735 pKa = 11.84EE736 pKa = 4.25AYY738 pKa = 9.4AVYY741 pKa = 9.88GYY743 pKa = 10.67SEE745 pKa = 5.35DD746 pKa = 3.9NGFSQHH752 pKa = 5.82MLEE755 pKa = 4.41EE756 pKa = 4.57TNGTGWVPTRR766 pKa = 11.84GIRR769 pKa = 11.84IGDD772 pKa = 3.33VLYY775 pKa = 10.79VVKK778 pKa = 10.85GNAAEE783 pKa = 4.25SYY785 pKa = 10.61RR786 pKa = 11.84IGDD789 pKa = 3.59YY790 pKa = 10.99QKK792 pKa = 10.6IDD794 pKa = 3.63DD795 pKa = 4.63LLII798 pKa = 4.67
Molecular weight: 84.02 kDa
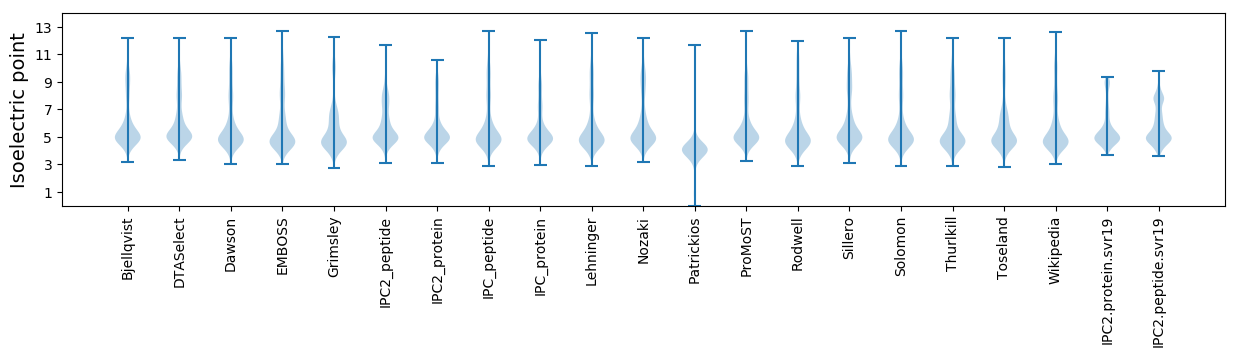
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N0ABY4|A0A3N0ABY4_9ACTN 10 kDa chaperonin OS=Adlercreutzia caecicola OX=747645 GN=groS PE=3 SV=1
MM1 pKa = 7.21TGGVGVIFLGVAVALAAVAGFGLAIGASALMRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LRR38 pKa = 11.84QTVATSAAAQGLGLSRR54 pKa = 11.84GASCVPGGARR64 pKa = 11.84AAPAVALLQAMEE76 pKa = 4.23LRR78 pKa = 11.84SRR80 pKa = 11.84ALSMGARR87 pKa = 11.84HH88 pKa = 6.25PLVPARR94 pKa = 11.84LLPAAASLEE103 pKa = 4.2KK104 pKa = 10.45AISHH108 pKa = 6.79AGLAGAVTVAGFWDD122 pKa = 4.36CSLLAALAGAVAGGLAGAAFSVEE145 pKa = 3.85LALIGLVAGGFAGFRR160 pKa = 11.84LPRR163 pKa = 11.84AVLARR168 pKa = 11.84AVQKK172 pKa = 10.73RR173 pKa = 11.84ADD175 pKa = 3.69DD176 pKa = 4.11LEE178 pKa = 5.74RR179 pKa = 11.84SLSEE183 pKa = 3.81MLEE186 pKa = 4.26VVALGLRR193 pKa = 11.84SGLSFEE199 pKa = 4.78GALALYY205 pKa = 7.49TAHH208 pKa = 6.94FEE210 pKa = 4.48CPLATGMAAAQAQWSCGLVSRR231 pKa = 11.84EE232 pKa = 3.56EE233 pKa = 4.01ALRR236 pKa = 11.84ALAASYY242 pKa = 10.74RR243 pKa = 11.84SLLFGRR249 pKa = 11.84VVEE252 pKa = 4.63TIVRR256 pKa = 11.84SIRR259 pKa = 11.84FGSSLADD266 pKa = 3.67NLDD269 pKa = 3.71DD270 pKa = 4.45AAAEE274 pKa = 4.01ARR276 pKa = 11.84AHH278 pKa = 5.8YY279 pKa = 7.96RR280 pKa = 11.84TARR283 pKa = 11.84QEE285 pKa = 4.4SVAKK289 pKa = 10.65APVKK293 pKa = 10.36MMLPTSTLILPAMLILVLGPVLLEE317 pKa = 4.14LMGG320 pKa = 4.35
MM1 pKa = 7.21TGGVGVIFLGVAVALAAVAGFGLAIGASALMRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LRR38 pKa = 11.84QTVATSAAAQGLGLSRR54 pKa = 11.84GASCVPGGARR64 pKa = 11.84AAPAVALLQAMEE76 pKa = 4.23LRR78 pKa = 11.84SRR80 pKa = 11.84ALSMGARR87 pKa = 11.84HH88 pKa = 6.25PLVPARR94 pKa = 11.84LLPAAASLEE103 pKa = 4.2KK104 pKa = 10.45AISHH108 pKa = 6.79AGLAGAVTVAGFWDD122 pKa = 4.36CSLLAALAGAVAGGLAGAAFSVEE145 pKa = 3.85LALIGLVAGGFAGFRR160 pKa = 11.84LPRR163 pKa = 11.84AVLARR168 pKa = 11.84AVQKK172 pKa = 10.73RR173 pKa = 11.84ADD175 pKa = 3.69DD176 pKa = 4.11LEE178 pKa = 5.74RR179 pKa = 11.84SLSEE183 pKa = 3.81MLEE186 pKa = 4.26VVALGLRR193 pKa = 11.84SGLSFEE199 pKa = 4.78GALALYY205 pKa = 7.49TAHH208 pKa = 6.94FEE210 pKa = 4.48CPLATGMAAAQAQWSCGLVSRR231 pKa = 11.84EE232 pKa = 3.56EE233 pKa = 4.01ALRR236 pKa = 11.84ALAASYY242 pKa = 10.74RR243 pKa = 11.84SLLFGRR249 pKa = 11.84VVEE252 pKa = 4.63TIVRR256 pKa = 11.84SIRR259 pKa = 11.84FGSSLADD266 pKa = 3.67NLDD269 pKa = 3.71DD270 pKa = 4.45AAAEE274 pKa = 4.01ARR276 pKa = 11.84AHH278 pKa = 5.8YY279 pKa = 7.96RR280 pKa = 11.84TARR283 pKa = 11.84QEE285 pKa = 4.4SVAKK289 pKa = 10.65APVKK293 pKa = 10.36MMLPTSTLILPAMLILVLGPVLLEE317 pKa = 4.14LMGG320 pKa = 4.35
Molecular weight: 32.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
688622 |
32 |
26543 |
378.2 |
41.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.038 ± 0.105 | 1.61 ± 0.065 |
5.705 ± 0.128 | 6.629 ± 0.056 |
3.81 ± 0.055 | 8.379 ± 0.063 |
1.663 ± 0.028 | 4.655 ± 0.085 |
3.724 ± 0.044 | 9.26 ± 0.143 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.052 | 3.148 ± 0.137 |
4.76 ± 0.043 | 3.514 ± 0.029 |
5.527 ± 0.089 | 5.575 ± 0.048 |
5.283 ± 0.157 | 7.97 ± 0.055 |
1.21 ± 0.03 | 2.906 ± 0.093 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
