
Sphingomonadales bacterium EhC05
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; unclassified Sphingomonadales
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
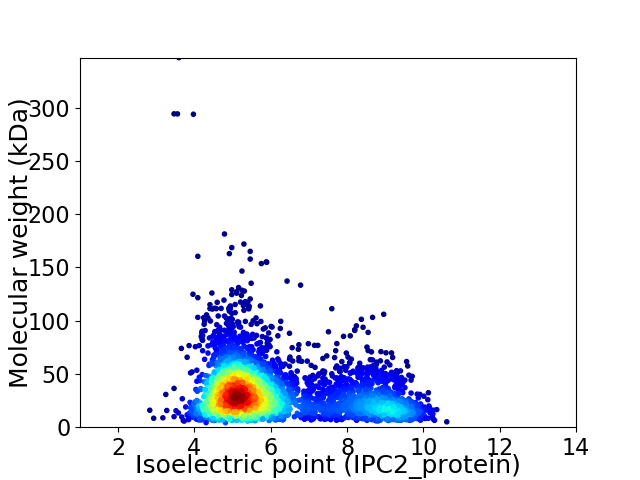
Virtual 2D-PAGE plot for 3899 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B6Z835|A0A1B6Z835_9SPHN Transport permease protein OS=Sphingomonadales bacterium EhC05 OX=1849171 GN=A8B75_16020 PE=3 SV=1
MM1 pKa = 7.79SGFLCLCLVALFPAEE16 pKa = 3.96KK17 pKa = 10.41AKK19 pKa = 10.94AQTITTYY26 pKa = 11.13SNSSTIAIPDD36 pKa = 3.69NACPATVNRR45 pKa = 11.84TFSVGSSYY53 pKa = 11.24EE54 pKa = 3.95VGDD57 pKa = 3.54VDD59 pKa = 5.01IGILLDD65 pKa = 3.15HH66 pKa = 7.06TYY68 pKa = 10.91RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.47LEE73 pKa = 4.2ITLTSPTGTIVTLMDD88 pKa = 3.95FVGGGANNLNVRR100 pKa = 11.84FDD102 pKa = 4.24DD103 pKa = 4.11EE104 pKa = 4.64ATGGTIASHH113 pKa = 6.36TSSDD117 pKa = 3.98PLTPIYY123 pKa = 10.74NSTKK127 pKa = 9.81IPQNSLSAFDD137 pKa = 5.38GEE139 pKa = 4.68DD140 pKa = 3.59AQGTWTLRR148 pKa = 11.84ICDD151 pKa = 3.52QQGIDD156 pKa = 3.27VGSFRR161 pKa = 11.84QADD164 pKa = 4.22LYY166 pKa = 9.88ITQLPPFADD175 pKa = 3.76LSLTKK180 pKa = 10.65SFGATSSNSGTYY192 pKa = 10.0SLSVTNSTISALTATGVTVSDD213 pKa = 3.95TLPSGVSLTGISGTGTYY230 pKa = 10.29SGGVWTLGTAIAPGQTVSIEE250 pKa = 3.95LSVTITATSGTITNSAQIRR269 pKa = 11.84SSTAFDD275 pKa = 4.08SDD277 pKa = 3.78STPNNGVTSEE287 pKa = 4.53DD288 pKa = 4.2DD289 pKa = 3.64YY290 pKa = 12.01ASVSFTAGTRR300 pKa = 11.84LPGYY304 pKa = 9.74VPPLTNVCPISNQLQFSWGSPTTWNPSGSLYY335 pKa = 9.1EE336 pKa = 4.37TYY338 pKa = 10.1TVTGIGEE345 pKa = 4.07IEE347 pKa = 4.05FTIAEE352 pKa = 4.28PPGGFTNSTPEE363 pKa = 3.64ITTSNTGGGAAGTRR377 pKa = 11.84ALYY380 pKa = 10.52FFMNNGNEE388 pKa = 4.28THH390 pKa = 6.71SSSTTLEE397 pKa = 4.33LPTAVPGLQFTIFDD411 pKa = 3.66IDD413 pKa = 3.67FGNNQFTDD421 pKa = 3.5MVTVTGEE428 pKa = 3.87FDD430 pKa = 3.37GATVLPTLSNNTANYY445 pKa = 9.75ISGNSAIGDD454 pKa = 3.81GSSSGTQEE462 pKa = 3.77FGNVVVTFTSPVDD475 pKa = 3.59SVTIVYY481 pKa = 10.54GNYY484 pKa = 9.63QPTTPTSSGNQAMSIHH500 pKa = 7.44DD501 pKa = 3.68FTFCKK506 pKa = 10.43PEE508 pKa = 3.86TTISITKK515 pKa = 10.19LSEE518 pKa = 3.91VLNDD522 pKa = 4.91PISPADD528 pKa = 3.1KK529 pKa = 9.93WKK531 pKa = 10.41RR532 pKa = 11.84IPDD535 pKa = 3.45ATVQYY540 pKa = 10.89CLFISNSGSGTAEE553 pKa = 3.71NVTVTDD559 pKa = 3.73NLVGPFTYY567 pKa = 9.75IAEE570 pKa = 4.38SMNSGANCDD579 pKa = 3.6SANTFEE585 pKa = 6.51DD586 pKa = 4.57DD587 pKa = 4.19DD588 pKa = 5.53SSDD591 pKa = 3.49GTEE594 pKa = 4.17TDD596 pKa = 5.38DD597 pKa = 3.54ITASIAGSTITVTAEE612 pKa = 4.3KK613 pKa = 10.13IGPTDD618 pKa = 3.25NFALTFRR625 pKa = 11.84VTIDD629 pKa = 2.85
MM1 pKa = 7.79SGFLCLCLVALFPAEE16 pKa = 3.96KK17 pKa = 10.41AKK19 pKa = 10.94AQTITTYY26 pKa = 11.13SNSSTIAIPDD36 pKa = 3.69NACPATVNRR45 pKa = 11.84TFSVGSSYY53 pKa = 11.24EE54 pKa = 3.95VGDD57 pKa = 3.54VDD59 pKa = 5.01IGILLDD65 pKa = 3.15HH66 pKa = 7.06TYY68 pKa = 10.91RR69 pKa = 11.84RR70 pKa = 11.84DD71 pKa = 3.47LEE73 pKa = 4.2ITLTSPTGTIVTLMDD88 pKa = 3.95FVGGGANNLNVRR100 pKa = 11.84FDD102 pKa = 4.24DD103 pKa = 4.11EE104 pKa = 4.64ATGGTIASHH113 pKa = 6.36TSSDD117 pKa = 3.98PLTPIYY123 pKa = 10.74NSTKK127 pKa = 9.81IPQNSLSAFDD137 pKa = 5.38GEE139 pKa = 4.68DD140 pKa = 3.59AQGTWTLRR148 pKa = 11.84ICDD151 pKa = 3.52QQGIDD156 pKa = 3.27VGSFRR161 pKa = 11.84QADD164 pKa = 4.22LYY166 pKa = 9.88ITQLPPFADD175 pKa = 3.76LSLTKK180 pKa = 10.65SFGATSSNSGTYY192 pKa = 10.0SLSVTNSTISALTATGVTVSDD213 pKa = 3.95TLPSGVSLTGISGTGTYY230 pKa = 10.29SGGVWTLGTAIAPGQTVSIEE250 pKa = 3.95LSVTITATSGTITNSAQIRR269 pKa = 11.84SSTAFDD275 pKa = 4.08SDD277 pKa = 3.78STPNNGVTSEE287 pKa = 4.53DD288 pKa = 4.2DD289 pKa = 3.64YY290 pKa = 12.01ASVSFTAGTRR300 pKa = 11.84LPGYY304 pKa = 9.74VPPLTNVCPISNQLQFSWGSPTTWNPSGSLYY335 pKa = 9.1EE336 pKa = 4.37TYY338 pKa = 10.1TVTGIGEE345 pKa = 4.07IEE347 pKa = 4.05FTIAEE352 pKa = 4.28PPGGFTNSTPEE363 pKa = 3.64ITTSNTGGGAAGTRR377 pKa = 11.84ALYY380 pKa = 10.52FFMNNGNEE388 pKa = 4.28THH390 pKa = 6.71SSSTTLEE397 pKa = 4.33LPTAVPGLQFTIFDD411 pKa = 3.66IDD413 pKa = 3.67FGNNQFTDD421 pKa = 3.5MVTVTGEE428 pKa = 3.87FDD430 pKa = 3.37GATVLPTLSNNTANYY445 pKa = 9.75ISGNSAIGDD454 pKa = 3.81GSSSGTQEE462 pKa = 3.77FGNVVVTFTSPVDD475 pKa = 3.59SVTIVYY481 pKa = 10.54GNYY484 pKa = 9.63QPTTPTSSGNQAMSIHH500 pKa = 7.44DD501 pKa = 3.68FTFCKK506 pKa = 10.43PEE508 pKa = 3.86TTISITKK515 pKa = 10.19LSEE518 pKa = 3.91VLNDD522 pKa = 4.91PISPADD528 pKa = 3.1KK529 pKa = 9.93WKK531 pKa = 10.41RR532 pKa = 11.84IPDD535 pKa = 3.45ATVQYY540 pKa = 10.89CLFISNSGSGTAEE553 pKa = 3.71NVTVTDD559 pKa = 3.73NLVGPFTYY567 pKa = 9.75IAEE570 pKa = 4.38SMNSGANCDD579 pKa = 3.6SANTFEE585 pKa = 6.51DD586 pKa = 4.57DD587 pKa = 4.19DD588 pKa = 5.53SSDD591 pKa = 3.49GTEE594 pKa = 4.17TDD596 pKa = 5.38DD597 pKa = 3.54ITASIAGSTITVTAEE612 pKa = 4.3KK613 pKa = 10.13IGPTDD618 pKa = 3.25NFALTFRR625 pKa = 11.84VTIDD629 pKa = 2.85
Molecular weight: 65.7 kDa
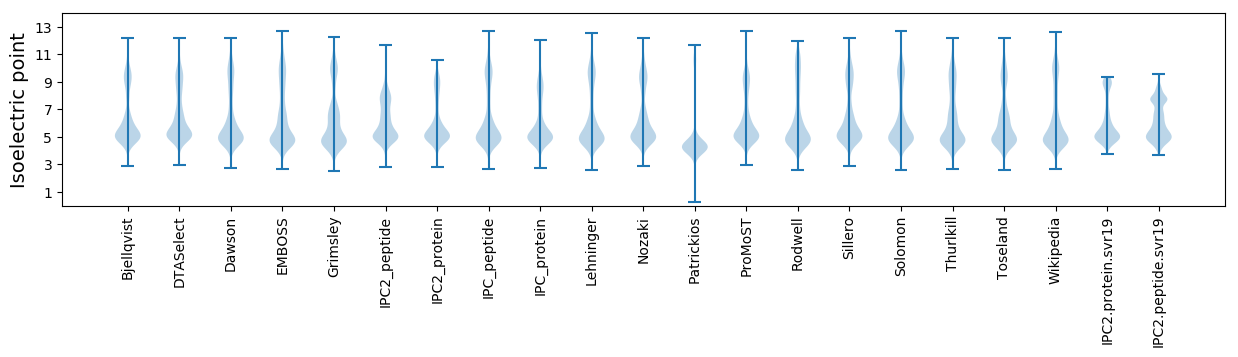
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B6Z791|A0A1B6Z791_9SPHN AB hydrolase-1 domain-containing protein OS=Sphingomonadales bacterium EhC05 OX=1849171 GN=A8B75_16870 PE=4 SV=1
MM1 pKa = 7.24TKK3 pKa = 9.49ATIAVSPLRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84MIDD17 pKa = 3.98DD18 pKa = 3.19MSLRR22 pKa = 11.84NLSPATQRR30 pKa = 11.84SYY32 pKa = 11.46LHH34 pKa = 6.74AVTKK38 pKa = 9.71FSRR41 pKa = 11.84FYY43 pKa = 11.2GRR45 pKa = 11.84SPDD48 pKa = 3.76CLGLEE53 pKa = 4.44DD54 pKa = 3.86VRR56 pKa = 11.84AFQVHH61 pKa = 6.25LVSQGISWGSLNQIVCALRR80 pKa = 11.84FFYY83 pKa = 10.57GVTLDD88 pKa = 3.41RR89 pKa = 11.84GEE91 pKa = 3.83IPKK94 pKa = 10.26RR95 pKa = 11.84IVYY98 pKa = 10.65ARR100 pKa = 11.84MPRR103 pKa = 11.84KK104 pKa = 10.02LPTILSADD112 pKa = 3.51EE113 pKa = 3.97VVRR116 pKa = 11.84FLEE119 pKa = 4.58AVPSLKK125 pKa = 10.44ARR127 pKa = 11.84AALTTAYY134 pKa = 10.36AAGLRR139 pKa = 11.84ASEE142 pKa = 4.32VASLRR147 pKa = 11.84VTDD150 pKa = 3.8IDD152 pKa = 3.85ADD154 pKa = 3.84RR155 pKa = 11.84MVIQIHH161 pKa = 6.13HH162 pKa = 6.67GKK164 pKa = 9.17VAQDD168 pKa = 3.37RR169 pKa = 11.84TVMLSPQLLAILRR182 pKa = 11.84TYY184 pKa = 9.07WRR186 pKa = 11.84LARR189 pKa = 11.84PPEE192 pKa = 4.14CLFPGRR198 pKa = 11.84TPSKK202 pKa = 9.9PIAITVLHH210 pKa = 6.34AACRR214 pKa = 11.84SATKK218 pKa = 10.36AAGLTKK224 pKa = 10.06RR225 pKa = 11.84VSVHH229 pKa = 4.88TLRR232 pKa = 11.84HH233 pKa = 5.12SFATHH238 pKa = 6.31LLEE241 pKa = 5.11SGVDD245 pKa = 3.19IRR247 pKa = 11.84IIQVLLGHH255 pKa = 6.84RR256 pKa = 11.84NLSTTALYY264 pKa = 8.74TQVATSTIAATQSPLDD280 pKa = 3.72RR281 pKa = 11.84LALEE285 pKa = 4.53VVPPTT290 pKa = 3.92
MM1 pKa = 7.24TKK3 pKa = 9.49ATIAVSPLRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84MIDD17 pKa = 3.98DD18 pKa = 3.19MSLRR22 pKa = 11.84NLSPATQRR30 pKa = 11.84SYY32 pKa = 11.46LHH34 pKa = 6.74AVTKK38 pKa = 9.71FSRR41 pKa = 11.84FYY43 pKa = 11.2GRR45 pKa = 11.84SPDD48 pKa = 3.76CLGLEE53 pKa = 4.44DD54 pKa = 3.86VRR56 pKa = 11.84AFQVHH61 pKa = 6.25LVSQGISWGSLNQIVCALRR80 pKa = 11.84FFYY83 pKa = 10.57GVTLDD88 pKa = 3.41RR89 pKa = 11.84GEE91 pKa = 3.83IPKK94 pKa = 10.26RR95 pKa = 11.84IVYY98 pKa = 10.65ARR100 pKa = 11.84MPRR103 pKa = 11.84KK104 pKa = 10.02LPTILSADD112 pKa = 3.51EE113 pKa = 3.97VVRR116 pKa = 11.84FLEE119 pKa = 4.58AVPSLKK125 pKa = 10.44ARR127 pKa = 11.84AALTTAYY134 pKa = 10.36AAGLRR139 pKa = 11.84ASEE142 pKa = 4.32VASLRR147 pKa = 11.84VTDD150 pKa = 3.8IDD152 pKa = 3.85ADD154 pKa = 3.84RR155 pKa = 11.84MVIQIHH161 pKa = 6.13HH162 pKa = 6.67GKK164 pKa = 9.17VAQDD168 pKa = 3.37RR169 pKa = 11.84TVMLSPQLLAILRR182 pKa = 11.84TYY184 pKa = 9.07WRR186 pKa = 11.84LARR189 pKa = 11.84PPEE192 pKa = 4.14CLFPGRR198 pKa = 11.84TPSKK202 pKa = 9.9PIAITVLHH210 pKa = 6.34AACRR214 pKa = 11.84SATKK218 pKa = 10.36AAGLTKK224 pKa = 10.06RR225 pKa = 11.84VSVHH229 pKa = 4.88TLRR232 pKa = 11.84HH233 pKa = 5.12SFATHH238 pKa = 6.31LLEE241 pKa = 5.11SGVDD245 pKa = 3.19IRR247 pKa = 11.84IIQVLLGHH255 pKa = 6.84RR256 pKa = 11.84NLSTTALYY264 pKa = 8.74TQVATSTIAATQSPLDD280 pKa = 3.72RR281 pKa = 11.84LALEE285 pKa = 4.53VVPPTT290 pKa = 3.92
Molecular weight: 32.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1226322 |
40 |
3577 |
314.5 |
34.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.072 ± 0.046 | 0.827 ± 0.013 |
6.27 ± 0.028 | 5.901 ± 0.038 |
3.943 ± 0.024 | 8.268 ± 0.052 |
1.956 ± 0.02 | 6.238 ± 0.027 |
4.207 ± 0.036 | 9.527 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.65 ± 0.022 | 3.42 ± 0.031 |
4.659 ± 0.028 | 3.556 ± 0.022 |
5.877 ± 0.038 | 6.199 ± 0.035 |
5.163 ± 0.031 | 6.486 ± 0.035 |
1.384 ± 0.016 | 2.396 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
