
Pichia membranifaciens NRRL Y-2026
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Pichiaceae; Pichia; Pichia membranifaciens
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
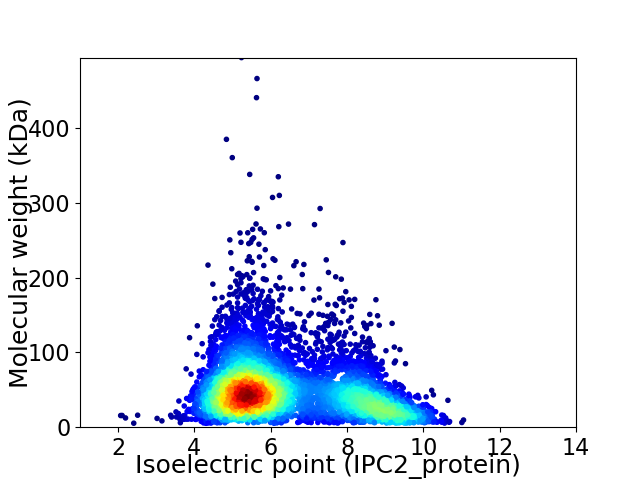
Virtual 2D-PAGE plot for 5536 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3NQ67|A0A1E3NQ67_9ASCO Uncharacterized protein OS=Pichia membranifaciens NRRL Y-2026 OX=763406 GN=PICMEDRAFT_125338 PE=4 SV=1
MM1 pKa = 6.97MSSYY5 pKa = 10.36FQSTYY10 pKa = 10.07NSGRR14 pKa = 11.84IAQTYY19 pKa = 10.23LLASKK24 pKa = 10.46VKK26 pKa = 10.81AKK28 pKa = 8.93LTQEE32 pKa = 3.95AVRR35 pKa = 11.84NDD37 pKa = 3.29VDD39 pKa = 3.6LHH41 pKa = 7.25RR42 pKa = 11.84LVCQANLLDD51 pKa = 4.82NLIEE55 pKa = 4.25NLNHH59 pKa = 6.78HH60 pKa = 6.23EE61 pKa = 4.06SSYY64 pKa = 11.62NSNTSSNPGKK74 pKa = 9.42TNVCYY79 pKa = 10.62DD80 pKa = 4.3SINSNSYY87 pKa = 9.8PLLNNVTVQVNDD99 pKa = 2.5VDD101 pKa = 4.22YY102 pKa = 11.48VVDD105 pKa = 3.98GDD107 pKa = 4.05ADD109 pKa = 3.7SGEE112 pKa = 4.28EE113 pKa = 4.14DD114 pKa = 4.92DD115 pKa = 5.88EE116 pKa = 4.84VDD118 pKa = 3.43EE119 pKa = 4.71NEE121 pKa = 4.29IKK123 pKa = 10.23IEE125 pKa = 3.94RR126 pKa = 11.84APCSDD131 pKa = 3.3VYY133 pKa = 10.22YY134 pKa = 10.53TSDD137 pKa = 4.79DD138 pKa = 3.7SDD140 pKa = 3.98FDD142 pKa = 4.97SDD144 pKa = 4.91DD145 pKa = 5.19DD146 pKa = 4.68SDD148 pKa = 4.64SSFNSDD154 pKa = 2.92TFGGKK159 pKa = 9.8SSDD162 pKa = 3.9PNEE165 pKa = 4.3LCCIALQRR173 pKa = 11.84LNLHH177 pKa = 6.38GGVEE181 pKa = 4.34DD182 pKa = 5.51GLGEE186 pKa = 5.6DD187 pKa = 5.88DD188 pKa = 6.6DD189 pKa = 5.8DD190 pKa = 6.23LEE192 pKa = 4.43NQATCSSVTATITLSGSDD210 pKa = 3.97SDD212 pKa = 4.72SEE214 pKa = 4.91SNSEE218 pKa = 4.18TEE220 pKa = 4.56SDD222 pKa = 3.27TEE224 pKa = 4.17YY225 pKa = 11.19EE226 pKa = 4.22NEE228 pKa = 4.07NEE230 pKa = 3.98NDD232 pKa = 3.78SRR234 pKa = 11.84DD235 pKa = 3.53HH236 pKa = 7.32GSFSNNYY243 pKa = 7.52CALMRR248 pKa = 11.84MHH250 pKa = 6.52SQHH253 pKa = 6.31TSLNLNDD260 pKa = 3.78YY261 pKa = 9.46VASLEE266 pKa = 4.07ADD268 pKa = 3.48TSVEE272 pKa = 3.98EE273 pKa = 4.43TDD275 pKa = 3.0ACTNIPHH282 pKa = 6.59NSSLDD287 pKa = 3.4EE288 pKa = 4.0MTRR291 pKa = 11.84EE292 pKa = 3.8EE293 pKa = 4.84DD294 pKa = 4.46LPSLSNCSSFSSMEE308 pKa = 4.25EE309 pKa = 3.2IGQPNQNIVRR319 pKa = 11.84LDD321 pKa = 3.55CLDD324 pKa = 4.36SIKK327 pKa = 10.45HH328 pKa = 4.63QHH330 pKa = 6.25DD331 pKa = 3.49TDD333 pKa = 4.06SPSLYY338 pKa = 9.2LTQSQDD344 pKa = 2.98VFLLL348 pKa = 3.98
MM1 pKa = 6.97MSSYY5 pKa = 10.36FQSTYY10 pKa = 10.07NSGRR14 pKa = 11.84IAQTYY19 pKa = 10.23LLASKK24 pKa = 10.46VKK26 pKa = 10.81AKK28 pKa = 8.93LTQEE32 pKa = 3.95AVRR35 pKa = 11.84NDD37 pKa = 3.29VDD39 pKa = 3.6LHH41 pKa = 7.25RR42 pKa = 11.84LVCQANLLDD51 pKa = 4.82NLIEE55 pKa = 4.25NLNHH59 pKa = 6.78HH60 pKa = 6.23EE61 pKa = 4.06SSYY64 pKa = 11.62NSNTSSNPGKK74 pKa = 9.42TNVCYY79 pKa = 10.62DD80 pKa = 4.3SINSNSYY87 pKa = 9.8PLLNNVTVQVNDD99 pKa = 2.5VDD101 pKa = 4.22YY102 pKa = 11.48VVDD105 pKa = 3.98GDD107 pKa = 4.05ADD109 pKa = 3.7SGEE112 pKa = 4.28EE113 pKa = 4.14DD114 pKa = 4.92DD115 pKa = 5.88EE116 pKa = 4.84VDD118 pKa = 3.43EE119 pKa = 4.71NEE121 pKa = 4.29IKK123 pKa = 10.23IEE125 pKa = 3.94RR126 pKa = 11.84APCSDD131 pKa = 3.3VYY133 pKa = 10.22YY134 pKa = 10.53TSDD137 pKa = 4.79DD138 pKa = 3.7SDD140 pKa = 3.98FDD142 pKa = 4.97SDD144 pKa = 4.91DD145 pKa = 5.19DD146 pKa = 4.68SDD148 pKa = 4.64SSFNSDD154 pKa = 2.92TFGGKK159 pKa = 9.8SSDD162 pKa = 3.9PNEE165 pKa = 4.3LCCIALQRR173 pKa = 11.84LNLHH177 pKa = 6.38GGVEE181 pKa = 4.34DD182 pKa = 5.51GLGEE186 pKa = 5.6DD187 pKa = 5.88DD188 pKa = 6.6DD189 pKa = 5.8DD190 pKa = 6.23LEE192 pKa = 4.43NQATCSSVTATITLSGSDD210 pKa = 3.97SDD212 pKa = 4.72SEE214 pKa = 4.91SNSEE218 pKa = 4.18TEE220 pKa = 4.56SDD222 pKa = 3.27TEE224 pKa = 4.17YY225 pKa = 11.19EE226 pKa = 4.22NEE228 pKa = 4.07NEE230 pKa = 3.98NDD232 pKa = 3.78SRR234 pKa = 11.84DD235 pKa = 3.53HH236 pKa = 7.32GSFSNNYY243 pKa = 7.52CALMRR248 pKa = 11.84MHH250 pKa = 6.52SQHH253 pKa = 6.31TSLNLNDD260 pKa = 3.78YY261 pKa = 9.46VASLEE266 pKa = 4.07ADD268 pKa = 3.48TSVEE272 pKa = 3.98EE273 pKa = 4.43TDD275 pKa = 3.0ACTNIPHH282 pKa = 6.59NSSLDD287 pKa = 3.4EE288 pKa = 4.0MTRR291 pKa = 11.84EE292 pKa = 3.8EE293 pKa = 4.84DD294 pKa = 4.46LPSLSNCSSFSSMEE308 pKa = 4.25EE309 pKa = 3.2IGQPNQNIVRR319 pKa = 11.84LDD321 pKa = 3.55CLDD324 pKa = 4.36SIKK327 pKa = 10.45HH328 pKa = 4.63QHH330 pKa = 6.25DD331 pKa = 3.49TDD333 pKa = 4.06SPSLYY338 pKa = 9.2LTQSQDD344 pKa = 2.98VFLLL348 pKa = 3.98
Molecular weight: 38.46 kDa
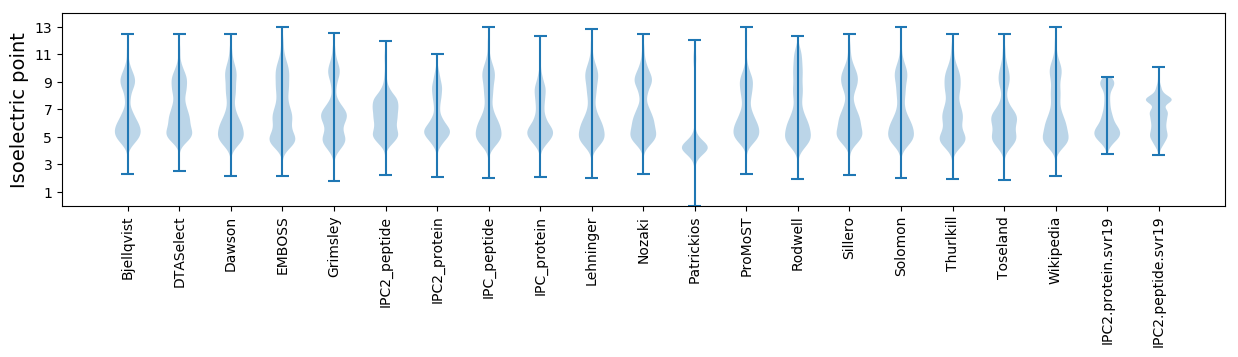
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3NHL2|A0A1E3NHL2_9ASCO Cysteinyl-tRNA synthetase OS=Pichia membranifaciens NRRL Y-2026 OX=763406 GN=PICMEDRAFT_16930 PE=3 SV=1
MM1 pKa = 7.93PSQKK5 pKa = 8.79TFRR8 pKa = 11.84VKK10 pKa = 10.56QKK12 pKa = 10.19LAKK15 pKa = 9.92AQNQNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNKK34 pKa = 8.41IRR36 pKa = 11.84YY37 pKa = 5.78NAKK40 pKa = 8.42RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.6WKK45 pKa = 8.07RR46 pKa = 11.84TKK48 pKa = 10.68LGLL51 pKa = 3.71
MM1 pKa = 7.93PSQKK5 pKa = 8.79TFRR8 pKa = 11.84VKK10 pKa = 10.56QKK12 pKa = 10.19LAKK15 pKa = 9.92AQNQNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNKK34 pKa = 8.41IRR36 pKa = 11.84YY37 pKa = 5.78NAKK40 pKa = 8.42RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.6WKK45 pKa = 8.07RR46 pKa = 11.84TKK48 pKa = 10.68LGLL51 pKa = 3.71
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2673506 |
49 |
4344 |
482.9 |
54.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.108 ± 0.037 | 1.284 ± 0.011 |
6.113 ± 0.028 | 6.572 ± 0.032 |
4.432 ± 0.024 | 5.463 ± 0.033 |
2.105 ± 0.014 | 6.15 ± 0.028 |
7.043 ± 0.03 | 9.533 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.111 ± 0.012 | 5.69 ± 0.03 |
4.282 ± 0.021 | 3.733 ± 0.029 |
4.408 ± 0.025 | 8.957 ± 0.044 |
5.555 ± 0.025 | 6.003 ± 0.024 |
1.017 ± 0.01 | 3.441 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
