
Aurantimonas sp. Leaf443
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aurantimonas; unclassified Aurantimonas
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
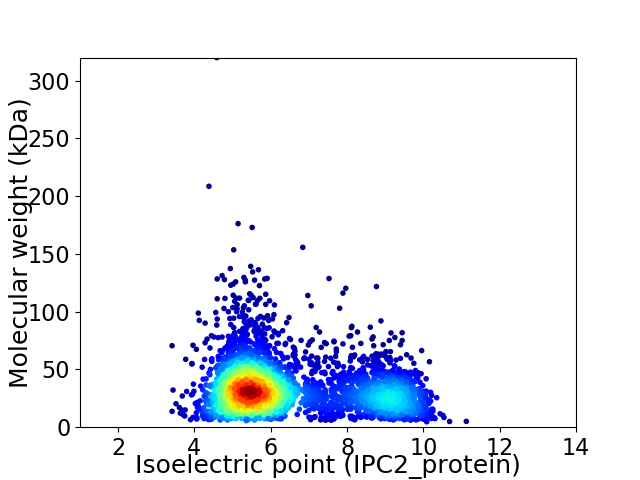
Virtual 2D-PAGE plot for 3439 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6F8A7|A0A0Q6F8A7_9RHIZ Cytochrome o ubiquinol oxidase subunit I OS=Aurantimonas sp. Leaf443 OX=1736378 GN=ASG48_17435 PE=3 SV=1
MM1 pKa = 7.41KK2 pKa = 10.47RR3 pKa = 11.84LALLLASAAFVTPALAADD21 pKa = 4.54VIYY24 pKa = 7.59DD25 pKa = 3.9TPVAPAAPMEE35 pKa = 4.31IAPVTEE41 pKa = 4.29TNTWTGLYY49 pKa = 9.84IGGQAGVAFGQDD61 pKa = 3.2SNDD64 pKa = 2.9IRR66 pKa = 11.84FDD68 pKa = 3.67PEE70 pKa = 3.65NNGAFGEE77 pKa = 4.76TIFEE81 pKa = 4.38NDD83 pKa = 3.18DD84 pKa = 3.49TNAGFVGGGHH94 pKa = 6.88IGYY97 pKa = 9.89DD98 pKa = 3.44YY99 pKa = 11.4QFGNNLVVGAVADD112 pKa = 4.13LNYY115 pKa = 10.06IDD117 pKa = 5.69AEE119 pKa = 4.32TSRR122 pKa = 11.84SYY124 pKa = 11.25SIDD127 pKa = 3.0GNRR130 pKa = 11.84FGVSEE135 pKa = 4.26DD136 pKa = 3.21LDD138 pKa = 3.87YY139 pKa = 11.4FGTVRR144 pKa = 11.84GRR146 pKa = 11.84LGYY149 pKa = 11.2AMDD152 pKa = 5.12SILVYY157 pKa = 10.79GSGGLAYY164 pKa = 10.35SGYY167 pKa = 10.56NRR169 pKa = 11.84DD170 pKa = 3.11NSFPSVPTNSLAGYY184 pKa = 6.41TFKK187 pKa = 11.12EE188 pKa = 4.59DD189 pKa = 3.61GDD191 pKa = 4.35DD192 pKa = 3.6VDD194 pKa = 3.82VGYY197 pKa = 10.86SVGGGIDD204 pKa = 3.48VMATRR209 pKa = 11.84NISFGVEE216 pKa = 3.57YY217 pKa = 10.34LYY219 pKa = 11.18TNLGKK224 pKa = 10.02NDD226 pKa = 3.67YY227 pKa = 10.69SVEE230 pKa = 4.0ATNGSNTIDD239 pKa = 3.49LTSTSKK245 pKa = 11.3DD246 pKa = 3.22DD247 pKa = 5.04LDD249 pKa = 3.78FHH251 pKa = 6.88TIFAKK256 pKa = 10.43ASYY259 pKa = 10.27RR260 pKa = 11.84FNN262 pKa = 3.56
MM1 pKa = 7.41KK2 pKa = 10.47RR3 pKa = 11.84LALLLASAAFVTPALAADD21 pKa = 4.54VIYY24 pKa = 7.59DD25 pKa = 3.9TPVAPAAPMEE35 pKa = 4.31IAPVTEE41 pKa = 4.29TNTWTGLYY49 pKa = 9.84IGGQAGVAFGQDD61 pKa = 3.2SNDD64 pKa = 2.9IRR66 pKa = 11.84FDD68 pKa = 3.67PEE70 pKa = 3.65NNGAFGEE77 pKa = 4.76TIFEE81 pKa = 4.38NDD83 pKa = 3.18DD84 pKa = 3.49TNAGFVGGGHH94 pKa = 6.88IGYY97 pKa = 9.89DD98 pKa = 3.44YY99 pKa = 11.4QFGNNLVVGAVADD112 pKa = 4.13LNYY115 pKa = 10.06IDD117 pKa = 5.69AEE119 pKa = 4.32TSRR122 pKa = 11.84SYY124 pKa = 11.25SIDD127 pKa = 3.0GNRR130 pKa = 11.84FGVSEE135 pKa = 4.26DD136 pKa = 3.21LDD138 pKa = 3.87YY139 pKa = 11.4FGTVRR144 pKa = 11.84GRR146 pKa = 11.84LGYY149 pKa = 11.2AMDD152 pKa = 5.12SILVYY157 pKa = 10.79GSGGLAYY164 pKa = 10.35SGYY167 pKa = 10.56NRR169 pKa = 11.84DD170 pKa = 3.11NSFPSVPTNSLAGYY184 pKa = 6.41TFKK187 pKa = 11.12EE188 pKa = 4.59DD189 pKa = 3.61GDD191 pKa = 4.35DD192 pKa = 3.6VDD194 pKa = 3.82VGYY197 pKa = 10.86SVGGGIDD204 pKa = 3.48VMATRR209 pKa = 11.84NISFGVEE216 pKa = 3.57YY217 pKa = 10.34LYY219 pKa = 11.18TNLGKK224 pKa = 10.02NDD226 pKa = 3.67YY227 pKa = 10.69SVEE230 pKa = 4.0ATNGSNTIDD239 pKa = 3.49LTSTSKK245 pKa = 11.3DD246 pKa = 3.22DD247 pKa = 5.04LDD249 pKa = 3.78FHH251 pKa = 6.88TIFAKK256 pKa = 10.43ASYY259 pKa = 10.27RR260 pKa = 11.84FNN262 pKa = 3.56
Molecular weight: 27.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6F141|A0A0Q6F141_9RHIZ GTPase HflX OS=Aurantimonas sp. Leaf443 OX=1736378 GN=hflX PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVIASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1115558 |
41 |
3002 |
324.4 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.026 ± 0.067 | 0.766 ± 0.012 |
5.569 ± 0.03 | 6.016 ± 0.036 |
3.732 ± 0.026 | 9.043 ± 0.044 |
1.841 ± 0.018 | 4.589 ± 0.032 |
2.621 ± 0.035 | 10.522 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.021 | 2.071 ± 0.02 |
5.36 ± 0.032 | 2.638 ± 0.023 |
7.867 ± 0.046 | 5.166 ± 0.031 |
5.164 ± 0.03 | 7.607 ± 0.035 |
1.109 ± 0.016 | 1.985 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
