
Streptococcus phage Javan261
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
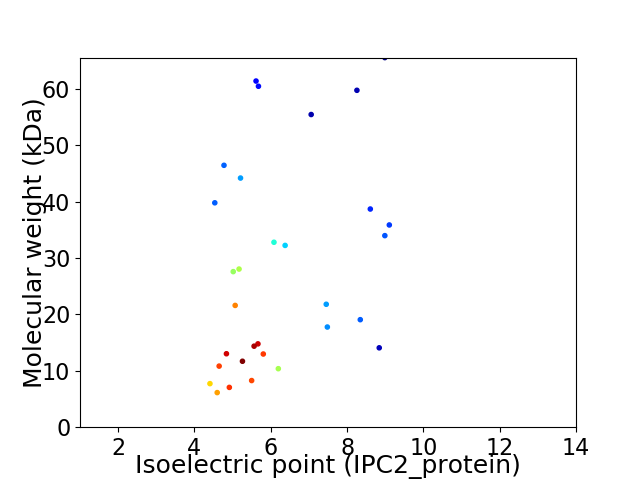
Virtual 2D-PAGE plot for 31 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6A6D8|A0A4D6A6D8_9CAUD Putative head-tail adaptor OS=Streptococcus phage Javan261 OX=2548078 GN=Javan261_0014 PE=4 SV=1
MM1 pKa = 7.47SGYY4 pKa = 8.89ATARR8 pKa = 11.84QNGWMSANDD17 pKa = 3.03IRR19 pKa = 11.84ALEE22 pKa = 4.08NLDD25 pKa = 5.16LISKK29 pKa = 10.14EE30 pKa = 4.26DD31 pKa = 3.62GGDD34 pKa = 3.24LYY36 pKa = 11.13LINGNMLPLKK46 pKa = 10.45DD47 pKa = 3.95AGAFAKK53 pKa = 10.34KK54 pKa = 10.29GEE56 pKa = 4.29HH57 pKa = 5.66NEE59 pKa = 4.03EE60 pKa = 4.09VLEE63 pKa = 5.17LEE65 pKa = 5.76DD66 pKa = 4.83DD67 pKa = 4.07TKK69 pKa = 11.21QQ70 pKa = 3.29
MM1 pKa = 7.47SGYY4 pKa = 8.89ATARR8 pKa = 11.84QNGWMSANDD17 pKa = 3.03IRR19 pKa = 11.84ALEE22 pKa = 4.08NLDD25 pKa = 5.16LISKK29 pKa = 10.14EE30 pKa = 4.26DD31 pKa = 3.62GGDD34 pKa = 3.24LYY36 pKa = 11.13LINGNMLPLKK46 pKa = 10.45DD47 pKa = 3.95AGAFAKK53 pKa = 10.34KK54 pKa = 10.29GEE56 pKa = 4.29HH57 pKa = 5.66NEE59 pKa = 4.03EE60 pKa = 4.09VLEE63 pKa = 5.17LEE65 pKa = 5.76DD66 pKa = 4.83DD67 pKa = 4.07TKK69 pKa = 11.21QQ70 pKa = 3.29
Molecular weight: 7.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6A6G4|A0A4D6A6G4_9CAUD Portal protein OS=Streptococcus phage Javan261 OX=2548078 GN=Javan261_0009 PE=4 SV=1
MM1 pKa = 6.96KK2 pKa = 10.13QIRR5 pKa = 11.84KK6 pKa = 9.35IIQIRR11 pKa = 11.84LPEE14 pKa = 4.02QKK16 pKa = 9.88KK17 pKa = 10.76LKK19 pKa = 8.17VAAYY23 pKa = 10.11ARR25 pKa = 11.84VSHH28 pKa = 5.99TRR30 pKa = 11.84LLQSLSNQVSYY41 pKa = 11.46YY42 pKa = 10.04NQIIGKK48 pKa = 8.38NPEE51 pKa = 3.7WEE53 pKa = 4.32FKK55 pKa = 10.44GVYY58 pKa = 8.84VDD60 pKa = 4.13EE61 pKa = 5.13VISGRR66 pKa = 11.84NASKK70 pKa = 10.2RR71 pKa = 11.84KK72 pKa = 9.0EE73 pKa = 4.03YY74 pKa = 10.53QRR76 pKa = 11.84LLSDD80 pKa = 3.59CRR82 pKa = 11.84KK83 pKa = 10.28GEE85 pKa = 3.84VDD87 pKa = 4.25IILTKK92 pKa = 10.5SISRR96 pKa = 11.84FGRR99 pKa = 11.84NTVEE103 pKa = 3.61LLKK106 pKa = 10.1TIRR109 pKa = 11.84EE110 pKa = 4.11LKK112 pKa = 9.63QLNISVRR119 pKa = 11.84FEE121 pKa = 4.05KK122 pKa = 10.93EE123 pKa = 3.82NIDD126 pKa = 3.69TLTTDD131 pKa = 3.91GEE133 pKa = 4.46LLLTLLSTLAEE144 pKa = 4.31EE145 pKa = 4.16EE146 pKa = 4.35SKK148 pKa = 11.09AISEE152 pKa = 4.2NIKK155 pKa = 10.18WKK157 pKa = 10.24VKK159 pKa = 10.21KK160 pKa = 10.27QFEE163 pKa = 4.11QGLPYY168 pKa = 10.1IKK170 pKa = 9.77QDD172 pKa = 2.99MYY174 pKa = 11.42GYY176 pKa = 10.21RR177 pKa = 11.84FKK179 pKa = 9.96EE180 pKa = 4.03TKK182 pKa = 10.55YY183 pKa = 10.16EE184 pKa = 3.94IEE186 pKa = 3.9PRR188 pKa = 11.84EE189 pKa = 4.17AEE191 pKa = 4.32VVKK194 pKa = 10.41QVYY197 pKa = 8.98AWYY200 pKa = 10.67LEE202 pKa = 4.5GEE204 pKa = 4.38QPTFIARR211 pKa = 11.84KK212 pKa = 9.72LNEE215 pKa = 3.91QGIKK219 pKa = 7.49TRR221 pKa = 11.84KK222 pKa = 8.5GNRR225 pKa = 11.84FSRR228 pKa = 11.84SIIHH232 pKa = 7.3RR233 pKa = 11.84ILGQEE238 pKa = 4.26AYY240 pKa = 9.36TGTLVLQKK248 pKa = 9.22TYY250 pKa = 10.85HH251 pKa = 5.96VGHH254 pKa = 7.31KK255 pKa = 10.11GRR257 pKa = 11.84SVEE260 pKa = 4.27NKK262 pKa = 10.21GEE264 pKa = 3.9RR265 pKa = 11.84TKK267 pKa = 11.34YY268 pKa = 9.51IVEE271 pKa = 4.24NAHH274 pKa = 5.61EE275 pKa = 4.68AIISRR280 pKa = 11.84EE281 pKa = 3.93MFEE284 pKa = 4.82EE285 pKa = 4.3VQQEE289 pKa = 4.0KK290 pKa = 10.69ARR292 pKa = 11.84RR293 pKa = 11.84SLHH296 pKa = 5.27NKK298 pKa = 8.77KK299 pKa = 10.44KK300 pKa = 10.27EE301 pKa = 3.98GRR303 pKa = 11.84HH304 pKa = 5.5DD305 pKa = 3.34
MM1 pKa = 6.96KK2 pKa = 10.13QIRR5 pKa = 11.84KK6 pKa = 9.35IIQIRR11 pKa = 11.84LPEE14 pKa = 4.02QKK16 pKa = 9.88KK17 pKa = 10.76LKK19 pKa = 8.17VAAYY23 pKa = 10.11ARR25 pKa = 11.84VSHH28 pKa = 5.99TRR30 pKa = 11.84LLQSLSNQVSYY41 pKa = 11.46YY42 pKa = 10.04NQIIGKK48 pKa = 8.38NPEE51 pKa = 3.7WEE53 pKa = 4.32FKK55 pKa = 10.44GVYY58 pKa = 8.84VDD60 pKa = 4.13EE61 pKa = 5.13VISGRR66 pKa = 11.84NASKK70 pKa = 10.2RR71 pKa = 11.84KK72 pKa = 9.0EE73 pKa = 4.03YY74 pKa = 10.53QRR76 pKa = 11.84LLSDD80 pKa = 3.59CRR82 pKa = 11.84KK83 pKa = 10.28GEE85 pKa = 3.84VDD87 pKa = 4.25IILTKK92 pKa = 10.5SISRR96 pKa = 11.84FGRR99 pKa = 11.84NTVEE103 pKa = 3.61LLKK106 pKa = 10.1TIRR109 pKa = 11.84EE110 pKa = 4.11LKK112 pKa = 9.63QLNISVRR119 pKa = 11.84FEE121 pKa = 4.05KK122 pKa = 10.93EE123 pKa = 3.82NIDD126 pKa = 3.69TLTTDD131 pKa = 3.91GEE133 pKa = 4.46LLLTLLSTLAEE144 pKa = 4.31EE145 pKa = 4.16EE146 pKa = 4.35SKK148 pKa = 11.09AISEE152 pKa = 4.2NIKK155 pKa = 10.18WKK157 pKa = 10.24VKK159 pKa = 10.21KK160 pKa = 10.27QFEE163 pKa = 4.11QGLPYY168 pKa = 10.1IKK170 pKa = 9.77QDD172 pKa = 2.99MYY174 pKa = 11.42GYY176 pKa = 10.21RR177 pKa = 11.84FKK179 pKa = 9.96EE180 pKa = 4.03TKK182 pKa = 10.55YY183 pKa = 10.16EE184 pKa = 3.94IEE186 pKa = 3.9PRR188 pKa = 11.84EE189 pKa = 4.17AEE191 pKa = 4.32VVKK194 pKa = 10.41QVYY197 pKa = 8.98AWYY200 pKa = 10.67LEE202 pKa = 4.5GEE204 pKa = 4.38QPTFIARR211 pKa = 11.84KK212 pKa = 9.72LNEE215 pKa = 3.91QGIKK219 pKa = 7.49TRR221 pKa = 11.84KK222 pKa = 8.5GNRR225 pKa = 11.84FSRR228 pKa = 11.84SIIHH232 pKa = 7.3RR233 pKa = 11.84ILGQEE238 pKa = 4.26AYY240 pKa = 9.36TGTLVLQKK248 pKa = 9.22TYY250 pKa = 10.85HH251 pKa = 5.96VGHH254 pKa = 7.31KK255 pKa = 10.11GRR257 pKa = 11.84SVEE260 pKa = 4.27NKK262 pKa = 10.21GEE264 pKa = 3.9RR265 pKa = 11.84TKK267 pKa = 11.34YY268 pKa = 9.51IVEE271 pKa = 4.24NAHH274 pKa = 5.61EE275 pKa = 4.68AIISRR280 pKa = 11.84EE281 pKa = 3.93MFEE284 pKa = 4.82EE285 pKa = 4.3VQQEE289 pKa = 4.0KK290 pKa = 10.69ARR292 pKa = 11.84RR293 pKa = 11.84SLHH296 pKa = 5.27NKK298 pKa = 8.77KK299 pKa = 10.44KK300 pKa = 10.27EE301 pKa = 3.98GRR303 pKa = 11.84HH304 pKa = 5.5DD305 pKa = 3.34
Molecular weight: 35.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7746 |
54 |
597 |
249.9 |
28.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.235 ± 0.504 | 0.981 ± 0.16 |
5.203 ± 0.448 | 7.449 ± 0.512 |
3.783 ± 0.237 | 6.791 ± 0.438 |
1.975 ± 0.205 | 6.894 ± 0.414 |
7.643 ± 0.506 | 8.817 ± 0.516 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.266 | 5.267 ± 0.218 |
2.969 ± 0.276 | 4.209 ± 0.203 |
4.299 ± 0.353 | 6.765 ± 0.421 |
6.636 ± 0.574 | 6.39 ± 0.293 |
1.33 ± 0.169 | 3.731 ± 0.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
