
Microcystis phage MACPNOA1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
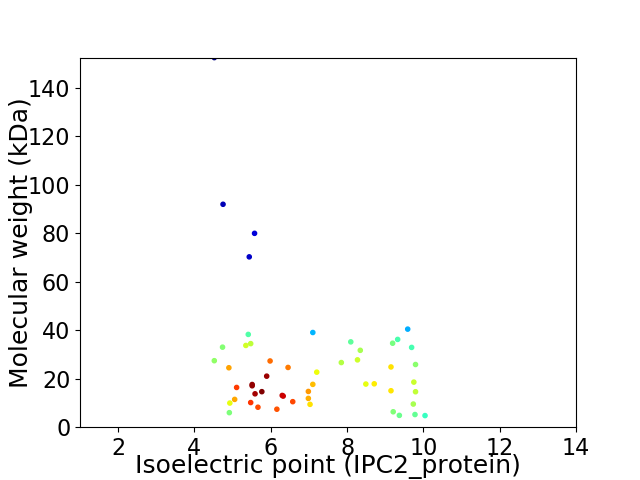
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3S6HCJ5|A0A3S6HCJ5_9CAUD Uncharacterized protein OS=Microcystis phage MACPNOA1 OX=1969741 PE=4 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84IALLLMLALLAGCASVPDD20 pKa = 4.61PNKK23 pKa = 10.09VDD25 pKa = 4.34PNHH28 pKa = 5.98QAYY31 pKa = 10.49LDD33 pKa = 3.61EE34 pKa = 4.5LAAIRR39 pKa = 11.84EE40 pKa = 4.39ANAQRR45 pKa = 11.84EE46 pKa = 4.32RR47 pKa = 11.84EE48 pKa = 3.98RR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 3.55AYY53 pKa = 10.43ARR55 pKa = 11.84MAEE58 pKa = 4.05ACAGDD63 pKa = 4.09AEE65 pKa = 4.66CVRR68 pKa = 11.84DD69 pKa = 3.88VARR72 pKa = 11.84EE73 pKa = 4.04ATLGEE78 pKa = 4.1AFANSGGRR86 pKa = 11.84RR87 pKa = 11.84DD88 pKa = 4.0APAPQPYY95 pKa = 9.36VRR97 pKa = 11.84PKK99 pKa = 9.62TKK101 pKa = 10.26GEE103 pKa = 3.89RR104 pKa = 11.84VAEE107 pKa = 3.94IVANNLTAGLVPSLVQGAVAVRR129 pKa = 11.84QSDD132 pKa = 3.48NSSAVAISGNEE143 pKa = 4.12MIRR146 pKa = 11.84DD147 pKa = 3.59IATGAIGVASQGLQQLPGLAPSYY170 pKa = 9.65TADD173 pKa = 3.44GDD175 pKa = 3.99IVLGDD180 pKa = 3.64NTTVGRR186 pKa = 11.84DD187 pKa = 3.07QWGSDD192 pKa = 3.0NRR194 pKa = 11.84VGDD197 pKa = 3.75EE198 pKa = 4.45FGDD201 pKa = 3.81INTGTQVEE209 pKa = 4.5SGIAGDD215 pKa = 4.44GNRR218 pKa = 11.84QASPDD223 pKa = 3.83DD224 pKa = 3.75YY225 pKa = 11.61SEE227 pKa = 4.16RR228 pKa = 11.84TEE230 pKa = 4.23CTGPGCQGTNRR241 pKa = 11.84PIRR244 pKa = 11.84IALPPEE250 pKa = 4.09RR251 pKa = 11.84PEE253 pKa = 4.01PEE255 pKa = 4.2EE256 pKa = 4.48PSDD259 pKa = 3.59EE260 pKa = 4.17
MM1 pKa = 7.92RR2 pKa = 11.84IALLLMLALLAGCASVPDD20 pKa = 4.61PNKK23 pKa = 10.09VDD25 pKa = 4.34PNHH28 pKa = 5.98QAYY31 pKa = 10.49LDD33 pKa = 3.61EE34 pKa = 4.5LAAIRR39 pKa = 11.84EE40 pKa = 4.39ANAQRR45 pKa = 11.84EE46 pKa = 4.32RR47 pKa = 11.84EE48 pKa = 3.98RR49 pKa = 11.84RR50 pKa = 11.84EE51 pKa = 3.55AYY53 pKa = 10.43ARR55 pKa = 11.84MAEE58 pKa = 4.05ACAGDD63 pKa = 4.09AEE65 pKa = 4.66CVRR68 pKa = 11.84DD69 pKa = 3.88VARR72 pKa = 11.84EE73 pKa = 4.04ATLGEE78 pKa = 4.1AFANSGGRR86 pKa = 11.84RR87 pKa = 11.84DD88 pKa = 4.0APAPQPYY95 pKa = 9.36VRR97 pKa = 11.84PKK99 pKa = 9.62TKK101 pKa = 10.26GEE103 pKa = 3.89RR104 pKa = 11.84VAEE107 pKa = 3.94IVANNLTAGLVPSLVQGAVAVRR129 pKa = 11.84QSDD132 pKa = 3.48NSSAVAISGNEE143 pKa = 4.12MIRR146 pKa = 11.84DD147 pKa = 3.59IATGAIGVASQGLQQLPGLAPSYY170 pKa = 9.65TADD173 pKa = 3.44GDD175 pKa = 3.99IVLGDD180 pKa = 3.64NTTVGRR186 pKa = 11.84DD187 pKa = 3.07QWGSDD192 pKa = 3.0NRR194 pKa = 11.84VGDD197 pKa = 3.75EE198 pKa = 4.45FGDD201 pKa = 3.81INTGTQVEE209 pKa = 4.5SGIAGDD215 pKa = 4.44GNRR218 pKa = 11.84QASPDD223 pKa = 3.83DD224 pKa = 3.75YY225 pKa = 11.61SEE227 pKa = 4.16RR228 pKa = 11.84TEE230 pKa = 4.23CTGPGCQGTNRR241 pKa = 11.84PIRR244 pKa = 11.84IALPPEE250 pKa = 4.09RR251 pKa = 11.84PEE253 pKa = 4.01PEE255 pKa = 4.2EE256 pKa = 4.48PSDD259 pKa = 3.59EE260 pKa = 4.17
Molecular weight: 27.42 kDa
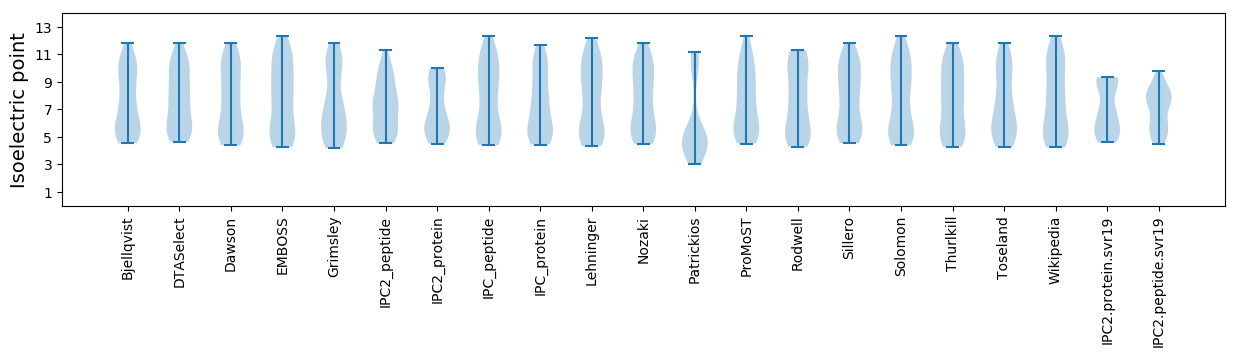
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S6HDH7|A0A3S6HDH7_9CAUD Uncharacterized protein OS=Microcystis phage MACPNOA1 OX=1969741 PE=4 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84ALSDD6 pKa = 3.27TASAVDD12 pKa = 3.43IEE14 pKa = 4.4AAIVHH19 pKa = 5.98HH20 pKa = 7.07LDD22 pKa = 2.92RR23 pKa = 11.84KK24 pKa = 8.96RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84GASANTLRR35 pKa = 11.84AYY37 pKa = 10.23AVDD40 pKa = 3.89LRR42 pKa = 11.84QFAAYY47 pKa = 9.5LVSRR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 3.9GVLAALVTHH62 pKa = 6.25RR63 pKa = 11.84HH64 pKa = 4.99IEE66 pKa = 3.95GWLDD70 pKa = 3.25TLSAQGVGARR80 pKa = 11.84SQARR84 pKa = 11.84KK85 pKa = 8.85LAVVRR90 pKa = 11.84GLFRR94 pKa = 11.84TCEE97 pKa = 3.85LEE99 pKa = 3.56GWIGHH104 pKa = 5.84DD105 pKa = 3.43PCRR108 pKa = 11.84DD109 pKa = 3.37VQVKK113 pKa = 8.19FRR115 pKa = 11.84RR116 pKa = 11.84VLRR119 pKa = 11.84TAPEE123 pKa = 4.16LPALYY128 pKa = 10.97AMVEE132 pKa = 4.63DD133 pKa = 4.46IGHH136 pKa = 4.77THH138 pKa = 6.0WRR140 pKa = 11.84DD141 pKa = 3.23LRR143 pKa = 11.84DD144 pKa = 3.38RR145 pKa = 11.84ALLRR149 pKa = 11.84LALDD153 pKa = 3.66AGLRR157 pKa = 11.84ISEE160 pKa = 4.46LLCLDD165 pKa = 3.51MAGCGPHH172 pKa = 6.39FVDD175 pKa = 4.47SKK177 pKa = 10.97RR178 pKa = 11.84LLVHH182 pKa = 6.51FGCKK186 pKa = 10.21GGDD189 pKa = 3.74SEE191 pKa = 4.97TSPINQRR198 pKa = 11.84TATIVQDD205 pKa = 4.15WIRR208 pKa = 11.84VRR210 pKa = 11.84GDD212 pKa = 2.92VAAPGEE218 pKa = 4.16TALFVSRR225 pKa = 11.84RR226 pKa = 11.84GTRR229 pKa = 11.84LTRR232 pKa = 11.84QMAHH236 pKa = 5.06VVVKK240 pKa = 10.02QRR242 pKa = 11.84GAAAGLPGLHH252 pKa = 5.64MHH254 pKa = 7.37LLRR257 pKa = 11.84HH258 pKa = 5.62RR259 pKa = 11.84RR260 pKa = 11.84IGDD263 pKa = 3.44VIEE266 pKa = 4.42RR267 pKa = 11.84CGLLAARR274 pKa = 11.84EE275 pKa = 4.17LAHH278 pKa = 6.72HH279 pKa = 6.74SSAEE283 pKa = 4.18TTGAVYY289 pKa = 10.25GRR291 pKa = 11.84HH292 pKa = 5.32VNTAAVQLLRR302 pKa = 11.84EE303 pKa = 4.15RR304 pKa = 11.84ADD306 pKa = 3.65LDD308 pKa = 3.88RR309 pKa = 11.84AGCAAA314 pKa = 4.47
MM1 pKa = 7.77RR2 pKa = 11.84ALSDD6 pKa = 3.27TASAVDD12 pKa = 3.43IEE14 pKa = 4.4AAIVHH19 pKa = 5.98HH20 pKa = 7.07LDD22 pKa = 2.92RR23 pKa = 11.84KK24 pKa = 8.96RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84GASANTLRR35 pKa = 11.84AYY37 pKa = 10.23AVDD40 pKa = 3.89LRR42 pKa = 11.84QFAAYY47 pKa = 9.5LVSRR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 3.9GVLAALVTHH62 pKa = 6.25RR63 pKa = 11.84HH64 pKa = 4.99IEE66 pKa = 3.95GWLDD70 pKa = 3.25TLSAQGVGARR80 pKa = 11.84SQARR84 pKa = 11.84KK85 pKa = 8.85LAVVRR90 pKa = 11.84GLFRR94 pKa = 11.84TCEE97 pKa = 3.85LEE99 pKa = 3.56GWIGHH104 pKa = 5.84DD105 pKa = 3.43PCRR108 pKa = 11.84DD109 pKa = 3.37VQVKK113 pKa = 8.19FRR115 pKa = 11.84RR116 pKa = 11.84VLRR119 pKa = 11.84TAPEE123 pKa = 4.16LPALYY128 pKa = 10.97AMVEE132 pKa = 4.63DD133 pKa = 4.46IGHH136 pKa = 4.77THH138 pKa = 6.0WRR140 pKa = 11.84DD141 pKa = 3.23LRR143 pKa = 11.84DD144 pKa = 3.38RR145 pKa = 11.84ALLRR149 pKa = 11.84LALDD153 pKa = 3.66AGLRR157 pKa = 11.84ISEE160 pKa = 4.46LLCLDD165 pKa = 3.51MAGCGPHH172 pKa = 6.39FVDD175 pKa = 4.47SKK177 pKa = 10.97RR178 pKa = 11.84LLVHH182 pKa = 6.51FGCKK186 pKa = 10.21GGDD189 pKa = 3.74SEE191 pKa = 4.97TSPINQRR198 pKa = 11.84TATIVQDD205 pKa = 4.15WIRR208 pKa = 11.84VRR210 pKa = 11.84GDD212 pKa = 2.92VAAPGEE218 pKa = 4.16TALFVSRR225 pKa = 11.84RR226 pKa = 11.84GTRR229 pKa = 11.84LTRR232 pKa = 11.84QMAHH236 pKa = 5.06VVVKK240 pKa = 10.02QRR242 pKa = 11.84GAAAGLPGLHH252 pKa = 5.64MHH254 pKa = 7.37LLRR257 pKa = 11.84HH258 pKa = 5.62RR259 pKa = 11.84RR260 pKa = 11.84IGDD263 pKa = 3.44VIEE266 pKa = 4.42RR267 pKa = 11.84CGLLAARR274 pKa = 11.84EE275 pKa = 4.17LAHH278 pKa = 6.72HH279 pKa = 6.74SSAEE283 pKa = 4.18TTGAVYY289 pKa = 10.25GRR291 pKa = 11.84HH292 pKa = 5.32VNTAAVQLLRR302 pKa = 11.84EE303 pKa = 4.15RR304 pKa = 11.84ADD306 pKa = 3.65LDD308 pKa = 3.88RR309 pKa = 11.84AGCAAA314 pKa = 4.47
Molecular weight: 34.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12656 |
45 |
1437 |
238.8 |
25.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.238 ± 0.564 | 1.075 ± 0.219 |
5.476 ± 0.196 | 6.479 ± 0.398 |
2.702 ± 0.194 | 8.454 ± 0.293 |
1.723 ± 0.289 | 3.848 ± 0.242 |
2.331 ± 0.273 | 9.956 ± 0.299 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.794 ± 0.126 | 2.347 ± 0.174 |
4.828 ± 0.361 | 4.772 ± 0.407 |
9.079 ± 0.354 | 5.136 ± 0.211 |
5.61 ± 0.407 | 6.606 ± 0.381 |
1.557 ± 0.206 | 1.991 ± 0.191 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
