
Marinoscillum sp. 108
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Reichenbachiellaceae; Marinoscillum; unclassified Marinoscillum
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
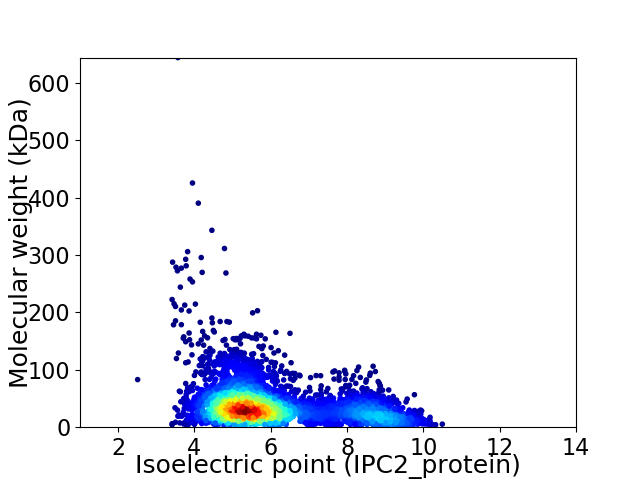
Virtual 2D-PAGE plot for 4475 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A654DYU8|A0A654DYU8_9BACT Uncharacterized protein OS=Marinoscillum sp. 108 OX=2653151 GN=MARINOS108_11871 PE=4 SV=1
MM1 pKa = 7.59LISSKK6 pKa = 10.14IYY8 pKa = 10.28RR9 pKa = 11.84FTFLLSLFSLLLTIACGGGDD29 pKa = 3.82GDD31 pKa = 6.11DD32 pKa = 3.98EE33 pKa = 4.79PEE35 pKa = 4.05PTAEE39 pKa = 4.8GILINEE45 pKa = 4.61IVASGDD51 pKa = 3.31DD52 pKa = 3.86WIEE55 pKa = 3.97LYY57 pKa = 10.7NALEE61 pKa = 4.12TSKK64 pKa = 10.92DD65 pKa = 3.42LSGYY69 pKa = 8.32TISDD73 pKa = 3.29SGNEE77 pKa = 4.04YY78 pKa = 10.33EE79 pKa = 5.34LPAGTTIAARR89 pKa = 11.84GFVVLLCNDD98 pKa = 4.25LGTGLNTNFKK108 pKa = 9.94LSSSGEE114 pKa = 4.28SVSLKK119 pKa = 10.15NAEE122 pKa = 4.05GTLIDD127 pKa = 3.97QVEE130 pKa = 4.67FPNLDD135 pKa = 3.23NGQSYY140 pKa = 11.4ARR142 pKa = 11.84FPDD145 pKa = 5.22GADD148 pKa = 2.68LWQITGVTTQGEE160 pKa = 5.04SNGSDD165 pKa = 3.3GAPAINSISRR175 pKa = 11.84SPLVPALNEE184 pKa = 4.2NVVVTAEE191 pKa = 4.64LISTTGVASVKK202 pKa = 10.21LFHH205 pKa = 7.05RR206 pKa = 11.84FNGSAFSEE214 pKa = 4.62VAMTLQSGTSYY225 pKa = 10.46TGTIPGMATEE235 pKa = 4.61GTVEE239 pKa = 4.29YY240 pKa = 10.4YY241 pKa = 10.57VEE243 pKa = 4.34AVGTNGSSSFKK254 pKa = 10.1PASAPDD260 pKa = 3.51NTDD263 pKa = 4.03DD264 pKa = 4.55YY265 pKa = 11.85LLNTDD270 pKa = 5.09PLPQLVINEE279 pKa = 3.99FMASNTSCCPDD290 pKa = 3.32TDD292 pKa = 3.77SGEE295 pKa = 4.83DD296 pKa = 4.07EE297 pKa = 4.21FDD299 pKa = 3.31DD300 pKa = 4.6WIEE303 pKa = 3.77IHH305 pKa = 6.47NLGATAVNLADD316 pKa = 4.67MYY318 pKa = 11.37FSDD321 pKa = 5.83DD322 pKa = 3.53KK323 pKa = 11.76DD324 pKa = 3.71NPFGDD329 pKa = 4.91KK330 pKa = 10.59ISDD333 pKa = 3.69DD334 pKa = 3.82DD335 pKa = 4.4AAATTIPAGGYY346 pKa = 9.7LVFWADD352 pKa = 3.36GSTSQGPTHH361 pKa = 6.93LNFSLSADD369 pKa = 3.79GEE371 pKa = 4.72DD372 pKa = 3.36VGLFYY377 pKa = 10.4IDD379 pKa = 3.81GRR381 pKa = 11.84TIDD384 pKa = 4.24AYY386 pKa = 9.9TFGAQSEE393 pKa = 4.45NVSWGRR399 pKa = 11.84ITDD402 pKa = 4.44GGDD405 pKa = 2.59TWGAQATPTPGQANQQ420 pKa = 3.17
MM1 pKa = 7.59LISSKK6 pKa = 10.14IYY8 pKa = 10.28RR9 pKa = 11.84FTFLLSLFSLLLTIACGGGDD29 pKa = 3.82GDD31 pKa = 6.11DD32 pKa = 3.98EE33 pKa = 4.79PEE35 pKa = 4.05PTAEE39 pKa = 4.8GILINEE45 pKa = 4.61IVASGDD51 pKa = 3.31DD52 pKa = 3.86WIEE55 pKa = 3.97LYY57 pKa = 10.7NALEE61 pKa = 4.12TSKK64 pKa = 10.92DD65 pKa = 3.42LSGYY69 pKa = 8.32TISDD73 pKa = 3.29SGNEE77 pKa = 4.04YY78 pKa = 10.33EE79 pKa = 5.34LPAGTTIAARR89 pKa = 11.84GFVVLLCNDD98 pKa = 4.25LGTGLNTNFKK108 pKa = 9.94LSSSGEE114 pKa = 4.28SVSLKK119 pKa = 10.15NAEE122 pKa = 4.05GTLIDD127 pKa = 3.97QVEE130 pKa = 4.67FPNLDD135 pKa = 3.23NGQSYY140 pKa = 11.4ARR142 pKa = 11.84FPDD145 pKa = 5.22GADD148 pKa = 2.68LWQITGVTTQGEE160 pKa = 5.04SNGSDD165 pKa = 3.3GAPAINSISRR175 pKa = 11.84SPLVPALNEE184 pKa = 4.2NVVVTAEE191 pKa = 4.64LISTTGVASVKK202 pKa = 10.21LFHH205 pKa = 7.05RR206 pKa = 11.84FNGSAFSEE214 pKa = 4.62VAMTLQSGTSYY225 pKa = 10.46TGTIPGMATEE235 pKa = 4.61GTVEE239 pKa = 4.29YY240 pKa = 10.4YY241 pKa = 10.57VEE243 pKa = 4.34AVGTNGSSSFKK254 pKa = 10.1PASAPDD260 pKa = 3.51NTDD263 pKa = 4.03DD264 pKa = 4.55YY265 pKa = 11.85LLNTDD270 pKa = 5.09PLPQLVINEE279 pKa = 3.99FMASNTSCCPDD290 pKa = 3.32TDD292 pKa = 3.77SGEE295 pKa = 4.83DD296 pKa = 4.07EE297 pKa = 4.21FDD299 pKa = 3.31DD300 pKa = 4.6WIEE303 pKa = 3.77IHH305 pKa = 6.47NLGATAVNLADD316 pKa = 4.67MYY318 pKa = 11.37FSDD321 pKa = 5.83DD322 pKa = 3.53KK323 pKa = 11.76DD324 pKa = 3.71NPFGDD329 pKa = 4.91KK330 pKa = 10.59ISDD333 pKa = 3.69DD334 pKa = 3.82DD335 pKa = 4.4AAATTIPAGGYY346 pKa = 9.7LVFWADD352 pKa = 3.36GSTSQGPTHH361 pKa = 6.93LNFSLSADD369 pKa = 3.79GEE371 pKa = 4.72DD372 pKa = 3.36VGLFYY377 pKa = 10.4IDD379 pKa = 3.81GRR381 pKa = 11.84TIDD384 pKa = 4.24AYY386 pKa = 9.9TFGAQSEE393 pKa = 4.45NVSWGRR399 pKa = 11.84ITDD402 pKa = 4.44GGDD405 pKa = 2.59TWGAQATPTPGQANQQ420 pKa = 3.17
Molecular weight: 44.36 kDa
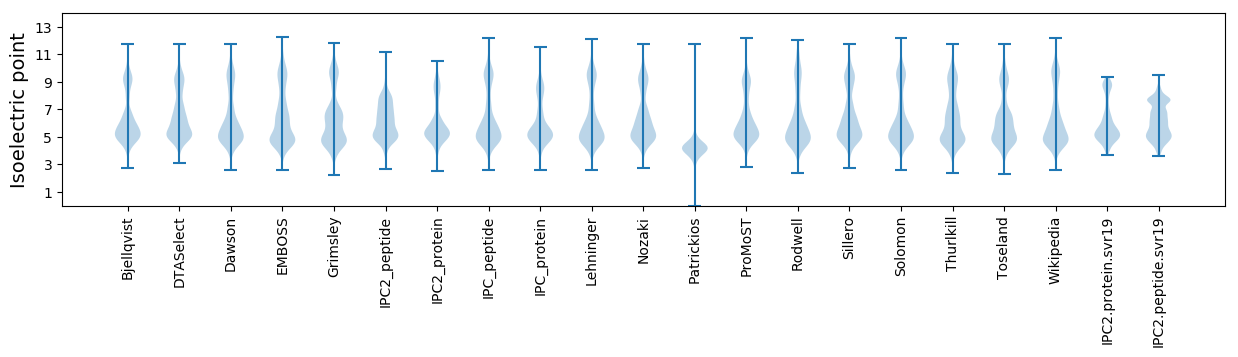
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A654E699|A0A654E699_9BACT DinB family protein OS=Marinoscillum sp. 108 OX=2653151 GN=MARINOS108_90125 PE=4 SV=1
MM1 pKa = 6.86IRR3 pKa = 11.84KK4 pKa = 9.33RR5 pKa = 11.84FITVVIRR12 pKa = 11.84IYY14 pKa = 11.06LNNLHH19 pKa = 6.67LEE21 pKa = 4.29KK22 pKa = 10.69EE23 pKa = 4.42QTTLTQTANSICHH36 pKa = 6.52LADD39 pKa = 3.19NLSYY43 pKa = 10.96LRR45 pKa = 11.84RR46 pKa = 11.84QKK48 pKa = 11.01GRR50 pKa = 11.84ICIKK54 pKa = 10.0IEE56 pKa = 3.85RR57 pKa = 11.84LNTIIPRR64 pKa = 11.84GYY66 pKa = 10.74
MM1 pKa = 6.86IRR3 pKa = 11.84KK4 pKa = 9.33RR5 pKa = 11.84FITVVIRR12 pKa = 11.84IYY14 pKa = 11.06LNNLHH19 pKa = 6.67LEE21 pKa = 4.29KK22 pKa = 10.69EE23 pKa = 4.42QTTLTQTANSICHH36 pKa = 6.52LADD39 pKa = 3.19NLSYY43 pKa = 10.96LRR45 pKa = 11.84RR46 pKa = 11.84QKK48 pKa = 11.01GRR50 pKa = 11.84ICIKK54 pKa = 10.0IEE56 pKa = 3.85RR57 pKa = 11.84LNTIIPRR64 pKa = 11.84GYY66 pKa = 10.74
Molecular weight: 7.86 kDa
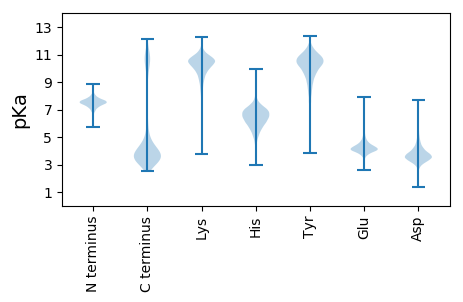
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1608423 |
24 |
6157 |
359.4 |
40.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.94 ± 0.032 | 0.692 ± 0.012 |
5.828 ± 0.035 | 6.62 ± 0.035 |
4.923 ± 0.027 | 7.247 ± 0.044 |
1.928 ± 0.019 | 6.915 ± 0.035 |
5.837 ± 0.052 | 9.558 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.379 ± 0.018 | 5.004 ± 0.034 |
3.756 ± 0.022 | 3.648 ± 0.021 |
4.053 ± 0.027 | 7.008 ± 0.045 |
5.788 ± 0.045 | 6.572 ± 0.026 |
1.25 ± 0.013 | 4.054 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
