
Puniceispirillum marinum (strain IMCC1322)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Alphaproteobacteria incertae sedis; SAR116 cluster; Candidatus Puniceispirillum; Candidatus Puniceispirillum marinum
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
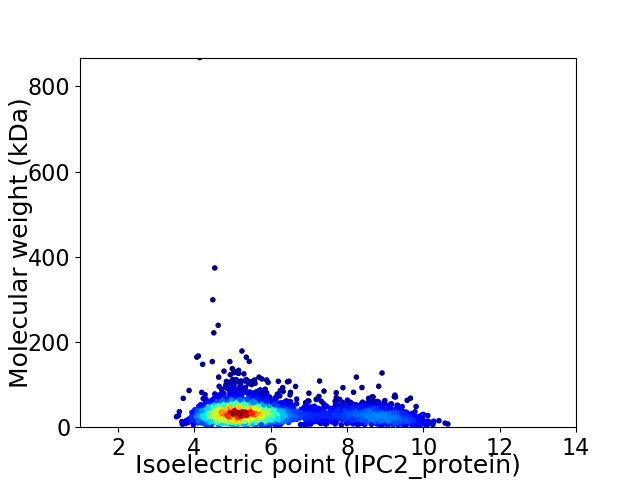
Virtual 2D-PAGE plot for 2545 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
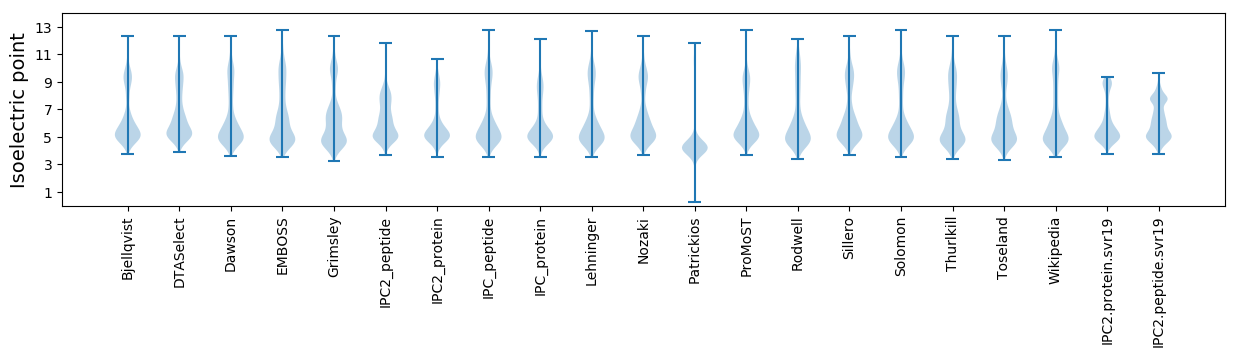
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5BQZ7|D5BQZ7_PUNMI Cation antiporter OS=Puniceispirillum marinum (strain IMCC1322) OX=488538 GN=SAR116_0467 PE=3 SV=1
MM1 pKa = 7.28VLNKK5 pKa = 9.53MSEE8 pKa = 4.44IISKK12 pKa = 7.94TANSEE17 pKa = 4.28TVTPSSMAGIFSLTEE32 pKa = 3.5AAADD36 pKa = 4.87RR37 pKa = 11.84ITSMLAAEE45 pKa = 4.55PDD47 pKa = 3.42GSFFRR52 pKa = 11.84VAVLGGGCSGFQYY65 pKa = 10.52QFSIDD70 pKa = 3.69TNRR73 pKa = 11.84DD74 pKa = 3.08DD75 pKa = 3.96EE76 pKa = 4.73DD77 pKa = 3.96QVFTSHH83 pKa = 6.23NVDD86 pKa = 3.72VIIDD90 pKa = 3.81EE91 pKa = 4.38MSLEE95 pKa = 4.25LVDD98 pKa = 4.98HH99 pKa = 7.19AEE101 pKa = 3.85LDD103 pKa = 4.08YY104 pKa = 11.61VQDD107 pKa = 3.64LMGSYY112 pKa = 10.21FAVNNPNATASCGCGTSFSVV132 pKa = 3.97
MM1 pKa = 7.28VLNKK5 pKa = 9.53MSEE8 pKa = 4.44IISKK12 pKa = 7.94TANSEE17 pKa = 4.28TVTPSSMAGIFSLTEE32 pKa = 3.5AAADD36 pKa = 4.87RR37 pKa = 11.84ITSMLAAEE45 pKa = 4.55PDD47 pKa = 3.42GSFFRR52 pKa = 11.84VAVLGGGCSGFQYY65 pKa = 10.52QFSIDD70 pKa = 3.69TNRR73 pKa = 11.84DD74 pKa = 3.08DD75 pKa = 3.96EE76 pKa = 4.73DD77 pKa = 3.96QVFTSHH83 pKa = 6.23NVDD86 pKa = 3.72VIIDD90 pKa = 3.81EE91 pKa = 4.38MSLEE95 pKa = 4.25LVDD98 pKa = 4.98HH99 pKa = 7.19AEE101 pKa = 3.85LDD103 pKa = 4.08YY104 pKa = 11.61VQDD107 pKa = 3.64LMGSYY112 pKa = 10.21FAVNNPNATASCGCGTSFSVV132 pKa = 3.97
Molecular weight: 14.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5BT02|D5BT02_PUNMI CDP-diacylglycerol--serine O-phosphatidyltransferase OS=Puniceispirillum marinum (strain IMCC1322) OX=488538 GN=SAR116_1156 PE=3 SV=1
MM1 pKa = 7.61FKK3 pKa = 10.68RR4 pKa = 11.84FIRR7 pKa = 11.84NKK9 pKa = 9.6FVIWCLSWVVAGIIASIMITTRR31 pKa = 11.84WKK33 pKa = 8.83TVNRR37 pKa = 11.84QLADD41 pKa = 3.48QILNNHH47 pKa = 6.18NGFVLVLWHH56 pKa = 6.07EE57 pKa = 4.81RR58 pKa = 11.84IFAVPWLWPRR68 pKa = 11.84RR69 pKa = 11.84HH70 pKa = 6.06PMYY73 pKa = 10.71VLQSPHH79 pKa = 7.08ADD81 pKa = 3.54GQLMSYY87 pKa = 8.11TVNRR91 pKa = 11.84MGIRR95 pKa = 11.84TVWGSSNRR103 pKa = 11.84NAISGLRR110 pKa = 11.84GLKK113 pKa = 9.83RR114 pKa = 11.84VLDD117 pKa = 4.19RR118 pKa = 11.84GQIATITPDD127 pKa = 3.45GPRR130 pKa = 11.84GPARR134 pKa = 11.84KK135 pKa = 8.74LAIGPVALAAMAGKK149 pKa = 9.15PVVPICWAVDD159 pKa = 3.98RR160 pKa = 11.84YY161 pKa = 8.84WRR163 pKa = 11.84APGWDD168 pKa = 3.32GTIIPKK174 pKa = 9.91PFARR178 pKa = 11.84GRR180 pKa = 11.84FIWGDD185 pKa = 3.69PIDD188 pKa = 4.11VPKK191 pKa = 10.82GDD193 pKa = 4.32RR194 pKa = 11.84DD195 pKa = 3.63TMEE198 pKa = 3.91TSRR201 pKa = 11.84VEE203 pKa = 3.99IEE205 pKa = 3.86TAMNDD210 pKa = 3.35LADD213 pKa = 4.08RR214 pKa = 11.84ADD216 pKa = 3.76RR217 pKa = 11.84LFAEE221 pKa = 4.62EE222 pKa = 4.77DD223 pKa = 3.61GLRR226 pKa = 3.71
MM1 pKa = 7.61FKK3 pKa = 10.68RR4 pKa = 11.84FIRR7 pKa = 11.84NKK9 pKa = 9.6FVIWCLSWVVAGIIASIMITTRR31 pKa = 11.84WKK33 pKa = 8.83TVNRR37 pKa = 11.84QLADD41 pKa = 3.48QILNNHH47 pKa = 6.18NGFVLVLWHH56 pKa = 6.07EE57 pKa = 4.81RR58 pKa = 11.84IFAVPWLWPRR68 pKa = 11.84RR69 pKa = 11.84HH70 pKa = 6.06PMYY73 pKa = 10.71VLQSPHH79 pKa = 7.08ADD81 pKa = 3.54GQLMSYY87 pKa = 8.11TVNRR91 pKa = 11.84MGIRR95 pKa = 11.84TVWGSSNRR103 pKa = 11.84NAISGLRR110 pKa = 11.84GLKK113 pKa = 9.83RR114 pKa = 11.84VLDD117 pKa = 4.19RR118 pKa = 11.84GQIATITPDD127 pKa = 3.45GPRR130 pKa = 11.84GPARR134 pKa = 11.84KK135 pKa = 8.74LAIGPVALAAMAGKK149 pKa = 9.15PVVPICWAVDD159 pKa = 3.98RR160 pKa = 11.84YY161 pKa = 8.84WRR163 pKa = 11.84APGWDD168 pKa = 3.32GTIIPKK174 pKa = 9.91PFARR178 pKa = 11.84GRR180 pKa = 11.84FIWGDD185 pKa = 3.69PIDD188 pKa = 4.11VPKK191 pKa = 10.82GDD193 pKa = 4.32RR194 pKa = 11.84DD195 pKa = 3.63TMEE198 pKa = 3.91TSRR201 pKa = 11.84VEE203 pKa = 3.99IEE205 pKa = 3.86TAMNDD210 pKa = 3.35LADD213 pKa = 4.08RR214 pKa = 11.84ADD216 pKa = 3.76RR217 pKa = 11.84LFAEE221 pKa = 4.62EE222 pKa = 4.77DD223 pKa = 3.61GLRR226 pKa = 3.71
Molecular weight: 25.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
832913 |
33 |
8246 |
327.3 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.876 ± 0.063 | 0.993 ± 0.02 |
6.609 ± 0.057 | 5.037 ± 0.042 |
3.993 ± 0.038 | 7.84 ± 0.046 |
2.329 ± 0.027 | 6.598 ± 0.044 |
4.153 ± 0.038 | 9.445 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.09 ± 0.035 | 3.59 ± 0.046 |
4.402 ± 0.031 | 3.363 ± 0.027 |
5.413 ± 0.041 | 6.167 ± 0.037 |
5.616 ± 0.071 | 6.82 ± 0.038 |
1.217 ± 0.02 | 2.451 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
