
Pseudoflavonifractor sp. BIOML-A4
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Pseudoflavonifractor; unclassified Pseudoflavonifractor
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
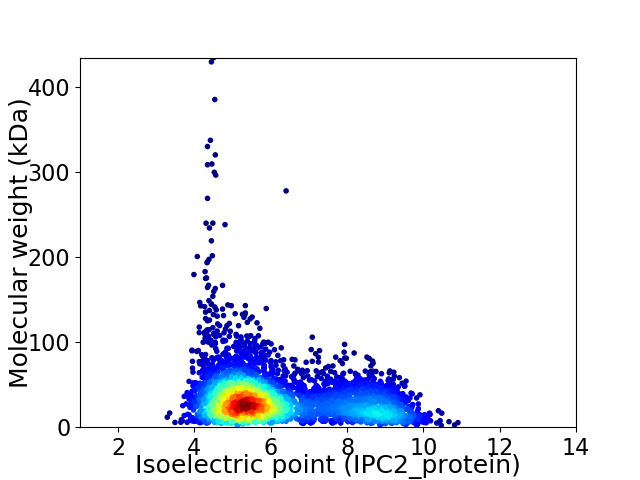
Virtual 2D-PAGE plot for 4105 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I3RAU3|A0A6I3RAU3_9FIRM Uncharacterized protein OS=Pseudoflavonifractor sp. BIOML-A4 OX=2584666 GN=GMD93_13595 PE=4 SV=1
MM1 pKa = 7.86LKK3 pKa = 10.35RR4 pKa = 11.84KK5 pKa = 9.56LIPLALTLGLLLSCAACGEE24 pKa = 4.61TAGGTASPSSTPSASPSSAPAAGEE48 pKa = 3.95TSIVLSDD55 pKa = 3.71GGVTVDD61 pKa = 4.26GAAASTDD68 pKa = 3.31AGSAVYY74 pKa = 10.16VGAEE78 pKa = 3.91IIYY81 pKa = 9.82YY82 pKa = 9.61EE83 pKa = 4.19AGRR86 pKa = 11.84DD87 pKa = 3.45EE88 pKa = 5.11SYY90 pKa = 11.41GAGAEE95 pKa = 4.28ADD97 pKa = 3.28GHH99 pKa = 5.18TAGEE103 pKa = 4.15AAEE106 pKa = 4.09QTVVTITQPGTYY118 pKa = 8.61RR119 pKa = 11.84VSGTLSAGQLAVDD132 pKa = 5.01LGEE135 pKa = 5.14DD136 pKa = 3.58AQSDD140 pKa = 3.69PDD142 pKa = 3.86AVVTLILDD150 pKa = 3.83NMDD153 pKa = 3.09ITCAVAPAVIFYY165 pKa = 10.66SVYY168 pKa = 9.85EE169 pKa = 4.43CGSADD174 pKa = 3.86TEE176 pKa = 4.45TTAATVDD183 pKa = 3.64TSAAGANVIIADD195 pKa = 3.76GSVNHH200 pKa = 6.24VDD202 pKa = 3.14GSYY205 pKa = 10.08VAKK208 pKa = 10.18IYY210 pKa = 10.67QEE212 pKa = 4.03GTEE215 pKa = 4.39KK216 pKa = 10.81KK217 pKa = 9.04LHH219 pKa = 6.42KK220 pKa = 10.22YY221 pKa = 10.51DD222 pKa = 3.19GAFYY226 pKa = 11.07SKK228 pKa = 10.46MSMNVQGEE236 pKa = 4.09AAGDD240 pKa = 3.8GEE242 pKa = 5.35LYY244 pKa = 8.86ITAANEE250 pKa = 4.05GLDD253 pKa = 3.95SEE255 pKa = 4.77LHH257 pKa = 5.07LTINGGNISITAQNDD272 pKa = 4.74GINTNEE278 pKa = 4.79DD279 pKa = 3.54GVSVTTVNGGRR290 pKa = 11.84LYY292 pKa = 10.65INAGLGDD299 pKa = 4.28EE300 pKa = 4.9GDD302 pKa = 4.78GIDD305 pKa = 3.54SNGYY309 pKa = 9.23LVINGGEE316 pKa = 4.03IVTLANGRR324 pKa = 11.84SGDD327 pKa = 3.61GGLDD331 pKa = 3.42ADD333 pKa = 4.47QGIYY337 pKa = 10.8LNGGSVLALGSRR349 pKa = 11.84NDD351 pKa = 3.6AASTDD356 pKa = 3.52SAQAYY361 pKa = 8.41MEE363 pKa = 4.54LAFSSTLPAGSVVSITDD380 pKa = 3.25AEE382 pKa = 4.47GNEE385 pKa = 4.15LVSYY389 pKa = 8.78TAEE392 pKa = 4.22KK393 pKa = 10.7EE394 pKa = 4.26FATLTYY400 pKa = 8.03TAPALRR406 pKa = 11.84DD407 pKa = 3.33DD408 pKa = 4.4AVYY411 pKa = 10.08HH412 pKa = 5.78VYY414 pKa = 10.77VDD416 pKa = 4.5GVQQQFTGHH425 pKa = 5.77SFGMGGGMMGGQEE438 pKa = 3.88PPEE441 pKa = 4.36GFEE444 pKa = 4.24HH445 pKa = 6.02GQEE448 pKa = 4.3GEE450 pKa = 4.2RR451 pKa = 11.84PDD453 pKa = 4.62LPEE456 pKa = 4.73GMEE459 pKa = 4.12PPEE462 pKa = 4.57DD463 pKa = 4.15FEE465 pKa = 6.28PGQQPDD471 pKa = 3.81GTGMPRR477 pKa = 11.84GTAPADD483 pKa = 4.11DD484 pKa = 4.86GDD486 pKa = 3.86RR487 pKa = 11.84TPPGGAPDD495 pKa = 3.76GSGRR499 pKa = 11.84GGGRR503 pKa = 11.84GDD505 pKa = 3.67RR506 pKa = 11.84GEE508 pKa = 4.24TLDD511 pKa = 3.99PSAEE515 pKa = 4.03FSLSAGSHH523 pKa = 5.4SFSGVGDD530 pKa = 3.78STLGG534 pKa = 3.34
MM1 pKa = 7.86LKK3 pKa = 10.35RR4 pKa = 11.84KK5 pKa = 9.56LIPLALTLGLLLSCAACGEE24 pKa = 4.61TAGGTASPSSTPSASPSSAPAAGEE48 pKa = 3.95TSIVLSDD55 pKa = 3.71GGVTVDD61 pKa = 4.26GAAASTDD68 pKa = 3.31AGSAVYY74 pKa = 10.16VGAEE78 pKa = 3.91IIYY81 pKa = 9.82YY82 pKa = 9.61EE83 pKa = 4.19AGRR86 pKa = 11.84DD87 pKa = 3.45EE88 pKa = 5.11SYY90 pKa = 11.41GAGAEE95 pKa = 4.28ADD97 pKa = 3.28GHH99 pKa = 5.18TAGEE103 pKa = 4.15AAEE106 pKa = 4.09QTVVTITQPGTYY118 pKa = 8.61RR119 pKa = 11.84VSGTLSAGQLAVDD132 pKa = 5.01LGEE135 pKa = 5.14DD136 pKa = 3.58AQSDD140 pKa = 3.69PDD142 pKa = 3.86AVVTLILDD150 pKa = 3.83NMDD153 pKa = 3.09ITCAVAPAVIFYY165 pKa = 10.66SVYY168 pKa = 9.85EE169 pKa = 4.43CGSADD174 pKa = 3.86TEE176 pKa = 4.45TTAATVDD183 pKa = 3.64TSAAGANVIIADD195 pKa = 3.76GSVNHH200 pKa = 6.24VDD202 pKa = 3.14GSYY205 pKa = 10.08VAKK208 pKa = 10.18IYY210 pKa = 10.67QEE212 pKa = 4.03GTEE215 pKa = 4.39KK216 pKa = 10.81KK217 pKa = 9.04LHH219 pKa = 6.42KK220 pKa = 10.22YY221 pKa = 10.51DD222 pKa = 3.19GAFYY226 pKa = 11.07SKK228 pKa = 10.46MSMNVQGEE236 pKa = 4.09AAGDD240 pKa = 3.8GEE242 pKa = 5.35LYY244 pKa = 8.86ITAANEE250 pKa = 4.05GLDD253 pKa = 3.95SEE255 pKa = 4.77LHH257 pKa = 5.07LTINGGNISITAQNDD272 pKa = 4.74GINTNEE278 pKa = 4.79DD279 pKa = 3.54GVSVTTVNGGRR290 pKa = 11.84LYY292 pKa = 10.65INAGLGDD299 pKa = 4.28EE300 pKa = 4.9GDD302 pKa = 4.78GIDD305 pKa = 3.54SNGYY309 pKa = 9.23LVINGGEE316 pKa = 4.03IVTLANGRR324 pKa = 11.84SGDD327 pKa = 3.61GGLDD331 pKa = 3.42ADD333 pKa = 4.47QGIYY337 pKa = 10.8LNGGSVLALGSRR349 pKa = 11.84NDD351 pKa = 3.6AASTDD356 pKa = 3.52SAQAYY361 pKa = 8.41MEE363 pKa = 4.54LAFSSTLPAGSVVSITDD380 pKa = 3.25AEE382 pKa = 4.47GNEE385 pKa = 4.15LVSYY389 pKa = 8.78TAEE392 pKa = 4.22KK393 pKa = 10.7EE394 pKa = 4.26FATLTYY400 pKa = 8.03TAPALRR406 pKa = 11.84DD407 pKa = 3.33DD408 pKa = 4.4AVYY411 pKa = 10.08HH412 pKa = 5.78VYY414 pKa = 10.77VDD416 pKa = 4.5GVQQQFTGHH425 pKa = 5.77SFGMGGGMMGGQEE438 pKa = 3.88PPEE441 pKa = 4.36GFEE444 pKa = 4.24HH445 pKa = 6.02GQEE448 pKa = 4.3GEE450 pKa = 4.2RR451 pKa = 11.84PDD453 pKa = 4.62LPEE456 pKa = 4.73GMEE459 pKa = 4.12PPEE462 pKa = 4.57DD463 pKa = 4.15FEE465 pKa = 6.28PGQQPDD471 pKa = 3.81GTGMPRR477 pKa = 11.84GTAPADD483 pKa = 4.11DD484 pKa = 4.86GDD486 pKa = 3.86RR487 pKa = 11.84TPPGGAPDD495 pKa = 3.76GSGRR499 pKa = 11.84GGGRR503 pKa = 11.84GDD505 pKa = 3.67RR506 pKa = 11.84GEE508 pKa = 4.24TLDD511 pKa = 3.99PSAEE515 pKa = 4.03FSLSAGSHH523 pKa = 5.4SFSGVGDD530 pKa = 3.78STLGG534 pKa = 3.34
Molecular weight: 53.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I3RCE7|A0A6I3RCE7_9FIRM DUF4314 domain-containing protein OS=Pseudoflavonifractor sp. BIOML-A4 OX=2584666 GN=GMD93_06560 PE=4 SV=1
SS1 pKa = 7.18RR2 pKa = 11.84GGGSFGGGGSRR13 pKa = 11.84GGGFGGGSRR22 pKa = 11.84GGGFGGRR29 pKa = 11.84RR30 pKa = 3.33
SS1 pKa = 7.18RR2 pKa = 11.84GGGSFGGGGSRR13 pKa = 11.84GGGFGGGSRR22 pKa = 11.84GGGFGGRR29 pKa = 11.84RR30 pKa = 3.33
Molecular weight: 2.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1302255 |
18 |
4012 |
317.2 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.197 ± 0.054 | 1.586 ± 0.02 |
5.7 ± 0.034 | 6.576 ± 0.039 |
3.822 ± 0.027 | 8.579 ± 0.047 |
1.687 ± 0.017 | 5.324 ± 0.037 |
4.411 ± 0.034 | 9.775 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.661 ± 0.02 | 3.335 ± 0.035 |
4.219 ± 0.03 | 3.001 ± 0.02 |
5.808 ± 0.055 | 5.605 ± 0.031 |
5.679 ± 0.05 | 7.322 ± 0.034 |
1.127 ± 0.017 | 3.587 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
