
Agarivorans albus MKT 106
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Agarivorans; Agarivorans albus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
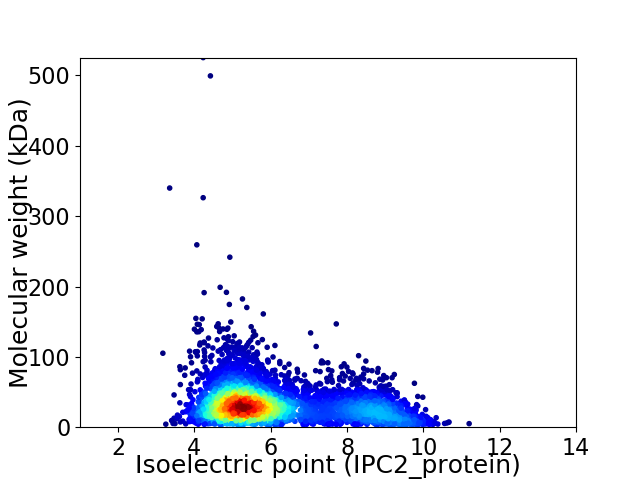
Virtual 2D-PAGE plot for 4397 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9PPL1|R9PPL1_AGAAL Putrescine utilization regulator OS=Agarivorans albus MKT 106 OX=1331007 GN=AALB_0141 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84LSRR5 pKa = 11.84IATAVTLFASVTIAGCGSSNSPFDD29 pKa = 3.98FGNDD33 pKa = 2.97ITPNEE38 pKa = 4.11PNSVKK43 pKa = 10.08IRR45 pKa = 11.84FNPLTGDD52 pKa = 3.06VSTPNNLLFNGSLDD66 pKa = 3.53GTINLPSEE74 pKa = 3.86AAIALTQQYY83 pKa = 10.78QDD85 pKa = 2.99ITNVLGALDD94 pKa = 4.39GWQTSVPFSIPLAYY108 pKa = 10.34GDD110 pKa = 5.64DD111 pKa = 3.43DD112 pKa = 4.8AEE114 pKa = 4.29YY115 pKa = 11.16NEE117 pKa = 5.19EE118 pKa = 4.51IDD120 pKa = 3.87TKK122 pKa = 11.15LDD124 pKa = 3.45ASSFPAGVLLYY135 pKa = 10.11KK136 pKa = 10.47VRR138 pKa = 11.84ASGFADD144 pKa = 3.58TCADD148 pKa = 3.32VTEE151 pKa = 4.67NGGSFVAGQTCRR163 pKa = 11.84VDD165 pKa = 3.25EE166 pKa = 3.96QLVYY170 pKa = 11.08GEE172 pKa = 5.35DD173 pKa = 4.05YY174 pKa = 10.85IATFSDD180 pKa = 3.58GSLIVTPLVPFAPATSYY197 pKa = 10.17MVAITDD203 pKa = 3.82QLLDD207 pKa = 3.6EE208 pKa = 4.95NGRR211 pKa = 11.84GVAAGEE217 pKa = 4.25AYY219 pKa = 10.11QLLSSPITADD229 pKa = 3.9DD230 pKa = 4.27DD231 pKa = 3.98ATVQSLKK238 pKa = 11.07GLTAFNNGLLAAEE251 pKa = 4.33GVTANVSYY259 pKa = 10.89SAVFTTQSVGSVLTTLQQIYY279 pKa = 10.82GGMALNSLLPLSALVDD295 pKa = 3.76TGTTAQDD302 pKa = 3.27VANAALGSSQPGFVAAKK319 pKa = 9.48YY320 pKa = 9.38FKK322 pKa = 9.98STLAVPYY329 pKa = 10.14YY330 pKa = 10.8LSDD333 pKa = 4.29LDD335 pKa = 5.09NPDD338 pKa = 3.1LTGADD343 pKa = 3.54VNKK346 pKa = 9.29WAQAKK351 pKa = 8.61TDD353 pKa = 3.66SPVAILQLLQTNADD367 pKa = 3.74FADD370 pKa = 3.64QGSATSFWAQAGAKK384 pKa = 9.29GFDD387 pKa = 3.18VATFGGLVAGGDD399 pKa = 3.76TTTAGQMLAQADD411 pKa = 4.38LAGLTYY417 pKa = 9.81MQDD420 pKa = 3.49GEE422 pKa = 4.35AMPVDD427 pKa = 3.77SHH429 pKa = 6.79RR430 pKa = 11.84HH431 pKa = 3.64LTAYY435 pKa = 10.72NPVPEE440 pKa = 4.5TKK442 pKa = 9.17STAFLDD448 pKa = 3.61LDD450 pKa = 3.75IYY452 pKa = 11.31LPDD455 pKa = 3.43EE456 pKa = 4.37TLTGNTMPAEE466 pKa = 4.49GWPVVIFQHH475 pKa = 7.08GIGSVKK481 pKa = 8.69EE482 pKa = 3.79TAAAIAASYY491 pKa = 9.21AAQGYY496 pKa = 9.62AVLAMDD502 pKa = 4.93LPYY505 pKa = 10.58HH506 pKa = 6.63GSRR509 pKa = 11.84GLDD512 pKa = 3.12LTGDD516 pKa = 3.88GNAEE520 pKa = 3.54ISANDD525 pKa = 3.81DD526 pKa = 3.42PQVFANLSSFLSIRR540 pKa = 11.84EE541 pKa = 3.88NLRR544 pKa = 11.84QSTADD549 pKa = 3.73LLAVRR554 pKa = 11.84AALQAAGAPAEE565 pKa = 4.01LDD567 pKa = 3.41GSKK570 pKa = 9.84VHH572 pKa = 6.53YY573 pKa = 10.35AGQSLGAIVGVGPASVADD591 pKa = 3.88TVNVSGFDD599 pKa = 3.08IDD601 pKa = 4.63YY602 pKa = 10.56SLQSVSLSAPAGGLAGVFQYY622 pKa = 10.86SPSFGPIVEE631 pKa = 5.23AGLKK635 pKa = 10.37SNPGYY640 pKa = 10.29LALVAAGLGFEE651 pKa = 5.53DD652 pKa = 5.15NDD654 pKa = 3.79TATALEE660 pKa = 4.13QFAVYY665 pKa = 9.75QATYY669 pKa = 10.13PEE671 pKa = 4.37AAQLVIDD678 pKa = 3.99SNYY681 pKa = 9.75PAFASIFGSLAQTVVDD697 pKa = 5.05GVDD700 pKa = 3.97PINYY704 pKa = 8.05AQSGFAMPVLVHH716 pKa = 6.63EE717 pKa = 4.53IVGDD721 pKa = 3.87GSEE724 pKa = 4.29GSGDD728 pKa = 3.51QVIPNSLATLPLVGTEE744 pKa = 4.01PLIASMGLEE753 pKa = 4.09GTEE756 pKa = 3.99VTSAGSYY763 pKa = 10.1AVRR766 pKa = 11.84FSQGSHH772 pKa = 6.06SSLISPAASLAATVEE787 pKa = 4.19MQTQFVSYY795 pKa = 10.24AVSADD800 pKa = 3.21AGNASVVVSDD810 pKa = 4.28SSVIAPAQQ818 pKa = 3.24
MM1 pKa = 7.62RR2 pKa = 11.84LSRR5 pKa = 11.84IATAVTLFASVTIAGCGSSNSPFDD29 pKa = 3.98FGNDD33 pKa = 2.97ITPNEE38 pKa = 4.11PNSVKK43 pKa = 10.08IRR45 pKa = 11.84FNPLTGDD52 pKa = 3.06VSTPNNLLFNGSLDD66 pKa = 3.53GTINLPSEE74 pKa = 3.86AAIALTQQYY83 pKa = 10.78QDD85 pKa = 2.99ITNVLGALDD94 pKa = 4.39GWQTSVPFSIPLAYY108 pKa = 10.34GDD110 pKa = 5.64DD111 pKa = 3.43DD112 pKa = 4.8AEE114 pKa = 4.29YY115 pKa = 11.16NEE117 pKa = 5.19EE118 pKa = 4.51IDD120 pKa = 3.87TKK122 pKa = 11.15LDD124 pKa = 3.45ASSFPAGVLLYY135 pKa = 10.11KK136 pKa = 10.47VRR138 pKa = 11.84ASGFADD144 pKa = 3.58TCADD148 pKa = 3.32VTEE151 pKa = 4.67NGGSFVAGQTCRR163 pKa = 11.84VDD165 pKa = 3.25EE166 pKa = 3.96QLVYY170 pKa = 11.08GEE172 pKa = 5.35DD173 pKa = 4.05YY174 pKa = 10.85IATFSDD180 pKa = 3.58GSLIVTPLVPFAPATSYY197 pKa = 10.17MVAITDD203 pKa = 3.82QLLDD207 pKa = 3.6EE208 pKa = 4.95NGRR211 pKa = 11.84GVAAGEE217 pKa = 4.25AYY219 pKa = 10.11QLLSSPITADD229 pKa = 3.9DD230 pKa = 4.27DD231 pKa = 3.98ATVQSLKK238 pKa = 11.07GLTAFNNGLLAAEE251 pKa = 4.33GVTANVSYY259 pKa = 10.89SAVFTTQSVGSVLTTLQQIYY279 pKa = 10.82GGMALNSLLPLSALVDD295 pKa = 3.76TGTTAQDD302 pKa = 3.27VANAALGSSQPGFVAAKK319 pKa = 9.48YY320 pKa = 9.38FKK322 pKa = 9.98STLAVPYY329 pKa = 10.14YY330 pKa = 10.8LSDD333 pKa = 4.29LDD335 pKa = 5.09NPDD338 pKa = 3.1LTGADD343 pKa = 3.54VNKK346 pKa = 9.29WAQAKK351 pKa = 8.61TDD353 pKa = 3.66SPVAILQLLQTNADD367 pKa = 3.74FADD370 pKa = 3.64QGSATSFWAQAGAKK384 pKa = 9.29GFDD387 pKa = 3.18VATFGGLVAGGDD399 pKa = 3.76TTTAGQMLAQADD411 pKa = 4.38LAGLTYY417 pKa = 9.81MQDD420 pKa = 3.49GEE422 pKa = 4.35AMPVDD427 pKa = 3.77SHH429 pKa = 6.79RR430 pKa = 11.84HH431 pKa = 3.64LTAYY435 pKa = 10.72NPVPEE440 pKa = 4.5TKK442 pKa = 9.17STAFLDD448 pKa = 3.61LDD450 pKa = 3.75IYY452 pKa = 11.31LPDD455 pKa = 3.43EE456 pKa = 4.37TLTGNTMPAEE466 pKa = 4.49GWPVVIFQHH475 pKa = 7.08GIGSVKK481 pKa = 8.69EE482 pKa = 3.79TAAAIAASYY491 pKa = 9.21AAQGYY496 pKa = 9.62AVLAMDD502 pKa = 4.93LPYY505 pKa = 10.58HH506 pKa = 6.63GSRR509 pKa = 11.84GLDD512 pKa = 3.12LTGDD516 pKa = 3.88GNAEE520 pKa = 3.54ISANDD525 pKa = 3.81DD526 pKa = 3.42PQVFANLSSFLSIRR540 pKa = 11.84EE541 pKa = 3.88NLRR544 pKa = 11.84QSTADD549 pKa = 3.73LLAVRR554 pKa = 11.84AALQAAGAPAEE565 pKa = 4.01LDD567 pKa = 3.41GSKK570 pKa = 9.84VHH572 pKa = 6.53YY573 pKa = 10.35AGQSLGAIVGVGPASVADD591 pKa = 3.88TVNVSGFDD599 pKa = 3.08IDD601 pKa = 4.63YY602 pKa = 10.56SLQSVSLSAPAGGLAGVFQYY622 pKa = 10.86SPSFGPIVEE631 pKa = 5.23AGLKK635 pKa = 10.37SNPGYY640 pKa = 10.29LALVAAGLGFEE651 pKa = 5.53DD652 pKa = 5.15NDD654 pKa = 3.79TATALEE660 pKa = 4.13QFAVYY665 pKa = 9.75QATYY669 pKa = 10.13PEE671 pKa = 4.37AAQLVIDD678 pKa = 3.99SNYY681 pKa = 9.75PAFASIFGSLAQTVVDD697 pKa = 5.05GVDD700 pKa = 3.97PINYY704 pKa = 8.05AQSGFAMPVLVHH716 pKa = 6.63EE717 pKa = 4.53IVGDD721 pKa = 3.87GSEE724 pKa = 4.29GSGDD728 pKa = 3.51QVIPNSLATLPLVGTEE744 pKa = 4.01PLIASMGLEE753 pKa = 4.09GTEE756 pKa = 3.99VTSAGSYY763 pKa = 10.1AVRR766 pKa = 11.84FSQGSHH772 pKa = 6.06SSLISPAASLAATVEE787 pKa = 4.19MQTQFVSYY795 pKa = 10.24AVSADD800 pKa = 3.21AGNASVVVSDD810 pKa = 4.28SSVIAPAQQ818 pKa = 3.24
Molecular weight: 84.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9PGZ8|R9PGZ8_AGAAL LemA protein OS=Agarivorans albus MKT 106 OX=1331007 GN=AALB_0697 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1381857 |
37 |
4824 |
314.3 |
34.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.308 ± 0.045 | 1.035 ± 0.015 |
5.325 ± 0.037 | 6.11 ± 0.035 |
4.096 ± 0.025 | 6.718 ± 0.036 |
2.181 ± 0.019 | 5.838 ± 0.028 |
4.937 ± 0.035 | 10.914 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.017 | 4.292 ± 0.028 |
3.873 ± 0.021 | 5.607 ± 0.04 |
4.275 ± 0.028 | 7.01 ± 0.039 |
4.722 ± 0.031 | 6.905 ± 0.029 |
1.354 ± 0.018 | 3.1 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
