
Psychroflexus salarius
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Psychroflexus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
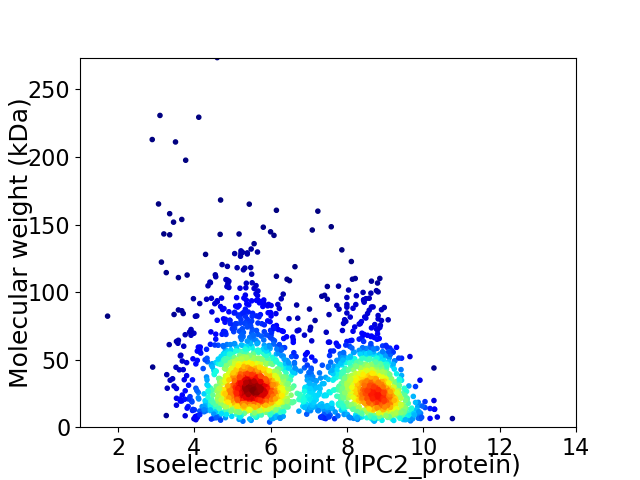
Virtual 2D-PAGE plot for 2311 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4XUK2|A0A1M4XUK2_9FLAO Glycosyltransferase catalytic subunit of cellulose synthase and poly-beta-1 6-N-acetylglucosamine synthase OS=Psychroflexus salarius OX=1155689 GN=SAMN05444278_11062 PE=4 SV=1
MM1 pKa = 7.69NYY3 pKa = 10.57NYY5 pKa = 10.71LLSCLLSLTLFSLGFSQSNPDD26 pKa = 3.56FYY28 pKa = 11.3LAEE31 pKa = 4.3NGVTCMCPDD40 pKa = 4.09ADD42 pKa = 4.8FGDD45 pKa = 4.32TGTLTINGVEE55 pKa = 3.97KK56 pKa = 10.26TFTKK60 pKa = 10.46RR61 pKa = 11.84SRR63 pKa = 11.84NLLYY67 pKa = 10.53AYY69 pKa = 9.7IDD71 pKa = 3.96VNPQDD76 pKa = 3.88PEE78 pKa = 3.84IALTCTSGITDD89 pKa = 4.52LSNAFNSKK97 pKa = 11.03SNFNQDD103 pKa = 2.82LSHH106 pKa = 6.89WDD108 pKa = 3.4VSNVTNMYY116 pKa = 10.94RR117 pKa = 11.84MFYY120 pKa = 9.33YY121 pKa = 10.87ANAFNQPLNNWDD133 pKa = 3.45VSNVTNMSDD142 pKa = 3.52MIYY145 pKa = 10.43SADD148 pKa = 4.17VFNQPLNNWDD158 pKa = 3.45VSNVTNMSNMFPGAIAFNQPLNNWDD183 pKa = 3.45VSNVTNMRR191 pKa = 11.84GMFYY195 pKa = 10.64GADD198 pKa = 3.38AFNQPLNNWDD208 pKa = 3.44VSSVTNLSVMFVDD221 pKa = 5.65ANAFNQPLDD230 pKa = 3.47NWDD233 pKa = 3.49VSSVTEE239 pKa = 3.97MNAMFTNATNFSQDD253 pKa = 3.02LSNWCVEE260 pKa = 4.84LITDD264 pKa = 5.02FPTSFASGSSMIPEE278 pKa = 4.76YY279 pKa = 10.97YY280 pKa = 9.5PDD282 pKa = 3.41WGAEE286 pKa = 4.02CSNAGTTTPVITLLGEE302 pKa = 4.04NPVTIEE308 pKa = 3.96VGTTYY313 pKa = 10.9TDD315 pKa = 3.35AGATASDD322 pKa = 3.88NTDD325 pKa = 2.8GDD327 pKa = 3.96LTDD330 pKa = 5.77DD331 pKa = 4.38IIVTGSVDD339 pKa = 3.21TDD341 pKa = 3.64VVGTYY346 pKa = 8.31TLSYY350 pKa = 11.06DD351 pKa = 3.73VTDD354 pKa = 3.66SSGNAAEE361 pKa = 4.41TVTRR365 pKa = 11.84TVNVVDD371 pKa = 3.41TTIPVITLLGDD382 pKa = 3.47NPVTIEE388 pKa = 4.02VGTTYY393 pKa = 10.9TDD395 pKa = 3.35AGATASDD402 pKa = 3.88NTDD405 pKa = 2.89GDD407 pKa = 3.83LTADD411 pKa = 4.13IIITGNVDD419 pKa = 3.08ADD421 pKa = 3.89VVGTYY426 pKa = 8.35TLSYY430 pKa = 11.06DD431 pKa = 3.73VTDD434 pKa = 3.66SSGNAAEE441 pKa = 4.24TVTRR445 pKa = 11.84IVNVVDD451 pKa = 3.15TTIPVISLLGDD462 pKa = 3.33NPVTIEE468 pKa = 3.99VSTTYY473 pKa = 10.66TDD475 pKa = 3.55AGATASDD482 pKa = 4.06NADD485 pKa = 3.12GDD487 pKa = 3.85LTANIIVTGSVDD499 pKa = 3.21TDD501 pKa = 3.64VVGTYY506 pKa = 8.31TLSYY510 pKa = 11.06DD511 pKa = 3.73VTDD514 pKa = 3.66SSGNAAEE521 pKa = 4.41TVTRR525 pKa = 11.84TVNVVGTTNPDD536 pKa = 3.23FYY538 pKa = 10.68IAEE541 pKa = 4.26NGVTCMCPDD550 pKa = 4.09ADD552 pKa = 4.8FGDD555 pKa = 4.32TGTLTINGEE564 pKa = 4.14EE565 pKa = 3.94KK566 pKa = 10.24TFTKK570 pKa = 9.83RR571 pKa = 11.84TRR573 pKa = 11.84AQLDD577 pKa = 3.73AIIANNQQDD586 pKa = 3.9PEE588 pKa = 4.15IALTCTSGITSMASLFSGKK607 pKa = 8.58DD608 pKa = 3.04TFNQDD613 pKa = 2.19ISHH616 pKa = 6.91WDD618 pKa = 3.32VSNVNNMQSAFIGASSFNQDD638 pKa = 1.91ISLWNTSNLVTMADD652 pKa = 3.58MFRR655 pKa = 11.84GASSFNNPLNNWDD668 pKa = 3.38VSNVVNMFSTFASASSFNQPLNNWDD693 pKa = 3.59VSSVTVMKK701 pKa = 10.88QMFASAEE708 pKa = 4.05AFNQSLDD715 pKa = 3.25NWDD718 pKa = 3.63VSNVTDD724 pKa = 3.61MSRR727 pKa = 11.84MFRR730 pKa = 11.84SAGSFNQTLNNWDD743 pKa = 3.53VSSVTNMNQMFAFADD758 pKa = 3.65AFNQSLNNWDD768 pKa = 3.62VSNVTDD774 pKa = 4.24MYY776 pKa = 11.61QMFNQAVTFNKK787 pKa = 9.82QLNDD791 pKa = 3.36WNTEE795 pKa = 3.42NVTNMEE801 pKa = 3.9GMFIIASSFNQPLDD815 pKa = 3.24QWDD818 pKa = 3.73VSAVNNMEE826 pKa = 4.48GMFKK830 pKa = 10.28QATSFNQPLNNWDD843 pKa = 3.49VSSVTNMRR851 pKa = 11.84NMFSGADD858 pKa = 3.44AFNQSLNNWNVSNVTDD874 pKa = 3.53MSFMFFGAVVFNQNISNWCVEE895 pKa = 4.42QIPSEE900 pKa = 3.99PSSFSTNSPLEE911 pKa = 3.98EE912 pKa = 4.0SFEE915 pKa = 4.8PNWGAEE921 pKa = 4.23CEE923 pKa = 4.36SLSIDD928 pKa = 3.98NPTEE932 pKa = 3.68AQLAIKK938 pKa = 9.83LYY940 pKa = 9.49PNPATHH946 pKa = 6.44QVTLTSNANADD957 pKa = 3.6VKK959 pKa = 10.96AVSLYY964 pKa = 11.4NMLGQKK970 pKa = 9.43VKK972 pKa = 10.54QLEE975 pKa = 4.34LSGSVAPIQIDD986 pKa = 3.83VSDD989 pKa = 4.57LAAGNYY995 pKa = 8.88FIQITTKK1002 pKa = 10.24NNTRR1006 pKa = 11.84ITKK1009 pKa = 10.21RR1010 pKa = 11.84FIKK1013 pKa = 10.39RR1014 pKa = 3.11
MM1 pKa = 7.69NYY3 pKa = 10.57NYY5 pKa = 10.71LLSCLLSLTLFSLGFSQSNPDD26 pKa = 3.56FYY28 pKa = 11.3LAEE31 pKa = 4.3NGVTCMCPDD40 pKa = 4.09ADD42 pKa = 4.8FGDD45 pKa = 4.32TGTLTINGVEE55 pKa = 3.97KK56 pKa = 10.26TFTKK60 pKa = 10.46RR61 pKa = 11.84SRR63 pKa = 11.84NLLYY67 pKa = 10.53AYY69 pKa = 9.7IDD71 pKa = 3.96VNPQDD76 pKa = 3.88PEE78 pKa = 3.84IALTCTSGITDD89 pKa = 4.52LSNAFNSKK97 pKa = 11.03SNFNQDD103 pKa = 2.82LSHH106 pKa = 6.89WDD108 pKa = 3.4VSNVTNMYY116 pKa = 10.94RR117 pKa = 11.84MFYY120 pKa = 9.33YY121 pKa = 10.87ANAFNQPLNNWDD133 pKa = 3.45VSNVTNMSDD142 pKa = 3.52MIYY145 pKa = 10.43SADD148 pKa = 4.17VFNQPLNNWDD158 pKa = 3.45VSNVTNMSNMFPGAIAFNQPLNNWDD183 pKa = 3.45VSNVTNMRR191 pKa = 11.84GMFYY195 pKa = 10.64GADD198 pKa = 3.38AFNQPLNNWDD208 pKa = 3.44VSSVTNLSVMFVDD221 pKa = 5.65ANAFNQPLDD230 pKa = 3.47NWDD233 pKa = 3.49VSSVTEE239 pKa = 3.97MNAMFTNATNFSQDD253 pKa = 3.02LSNWCVEE260 pKa = 4.84LITDD264 pKa = 5.02FPTSFASGSSMIPEE278 pKa = 4.76YY279 pKa = 10.97YY280 pKa = 9.5PDD282 pKa = 3.41WGAEE286 pKa = 4.02CSNAGTTTPVITLLGEE302 pKa = 4.04NPVTIEE308 pKa = 3.96VGTTYY313 pKa = 10.9TDD315 pKa = 3.35AGATASDD322 pKa = 3.88NTDD325 pKa = 2.8GDD327 pKa = 3.96LTDD330 pKa = 5.77DD331 pKa = 4.38IIVTGSVDD339 pKa = 3.21TDD341 pKa = 3.64VVGTYY346 pKa = 8.31TLSYY350 pKa = 11.06DD351 pKa = 3.73VTDD354 pKa = 3.66SSGNAAEE361 pKa = 4.41TVTRR365 pKa = 11.84TVNVVDD371 pKa = 3.41TTIPVITLLGDD382 pKa = 3.47NPVTIEE388 pKa = 4.02VGTTYY393 pKa = 10.9TDD395 pKa = 3.35AGATASDD402 pKa = 3.88NTDD405 pKa = 2.89GDD407 pKa = 3.83LTADD411 pKa = 4.13IIITGNVDD419 pKa = 3.08ADD421 pKa = 3.89VVGTYY426 pKa = 8.35TLSYY430 pKa = 11.06DD431 pKa = 3.73VTDD434 pKa = 3.66SSGNAAEE441 pKa = 4.24TVTRR445 pKa = 11.84IVNVVDD451 pKa = 3.15TTIPVISLLGDD462 pKa = 3.33NPVTIEE468 pKa = 3.99VSTTYY473 pKa = 10.66TDD475 pKa = 3.55AGATASDD482 pKa = 4.06NADD485 pKa = 3.12GDD487 pKa = 3.85LTANIIVTGSVDD499 pKa = 3.21TDD501 pKa = 3.64VVGTYY506 pKa = 8.31TLSYY510 pKa = 11.06DD511 pKa = 3.73VTDD514 pKa = 3.66SSGNAAEE521 pKa = 4.41TVTRR525 pKa = 11.84TVNVVGTTNPDD536 pKa = 3.23FYY538 pKa = 10.68IAEE541 pKa = 4.26NGVTCMCPDD550 pKa = 4.09ADD552 pKa = 4.8FGDD555 pKa = 4.32TGTLTINGEE564 pKa = 4.14EE565 pKa = 3.94KK566 pKa = 10.24TFTKK570 pKa = 9.83RR571 pKa = 11.84TRR573 pKa = 11.84AQLDD577 pKa = 3.73AIIANNQQDD586 pKa = 3.9PEE588 pKa = 4.15IALTCTSGITSMASLFSGKK607 pKa = 8.58DD608 pKa = 3.04TFNQDD613 pKa = 2.19ISHH616 pKa = 6.91WDD618 pKa = 3.32VSNVNNMQSAFIGASSFNQDD638 pKa = 1.91ISLWNTSNLVTMADD652 pKa = 3.58MFRR655 pKa = 11.84GASSFNNPLNNWDD668 pKa = 3.38VSNVVNMFSTFASASSFNQPLNNWDD693 pKa = 3.59VSSVTVMKK701 pKa = 10.88QMFASAEE708 pKa = 4.05AFNQSLDD715 pKa = 3.25NWDD718 pKa = 3.63VSNVTDD724 pKa = 3.61MSRR727 pKa = 11.84MFRR730 pKa = 11.84SAGSFNQTLNNWDD743 pKa = 3.53VSSVTNMNQMFAFADD758 pKa = 3.65AFNQSLNNWDD768 pKa = 3.62VSNVTDD774 pKa = 4.24MYY776 pKa = 11.61QMFNQAVTFNKK787 pKa = 9.82QLNDD791 pKa = 3.36WNTEE795 pKa = 3.42NVTNMEE801 pKa = 3.9GMFIIASSFNQPLDD815 pKa = 3.24QWDD818 pKa = 3.73VSAVNNMEE826 pKa = 4.48GMFKK830 pKa = 10.28QATSFNQPLNNWDD843 pKa = 3.49VSSVTNMRR851 pKa = 11.84NMFSGADD858 pKa = 3.44AFNQSLNNWNVSNVTDD874 pKa = 3.53MSFMFFGAVVFNQNISNWCVEE895 pKa = 4.42QIPSEE900 pKa = 3.99PSSFSTNSPLEE911 pKa = 3.98EE912 pKa = 4.0SFEE915 pKa = 4.8PNWGAEE921 pKa = 4.23CEE923 pKa = 4.36SLSIDD928 pKa = 3.98NPTEE932 pKa = 3.68AQLAIKK938 pKa = 9.83LYY940 pKa = 9.49PNPATHH946 pKa = 6.44QVTLTSNANADD957 pKa = 3.6VKK959 pKa = 10.96AVSLYY964 pKa = 11.4NMLGQKK970 pKa = 9.43VKK972 pKa = 10.54QLEE975 pKa = 4.34LSGSVAPIQIDD986 pKa = 3.83VSDD989 pKa = 4.57LAAGNYY995 pKa = 8.88FIQITTKK1002 pKa = 10.24NNTRR1006 pKa = 11.84ITKK1009 pKa = 10.21RR1010 pKa = 11.84FIKK1013 pKa = 10.39RR1014 pKa = 3.11
Molecular weight: 110.72 kDa
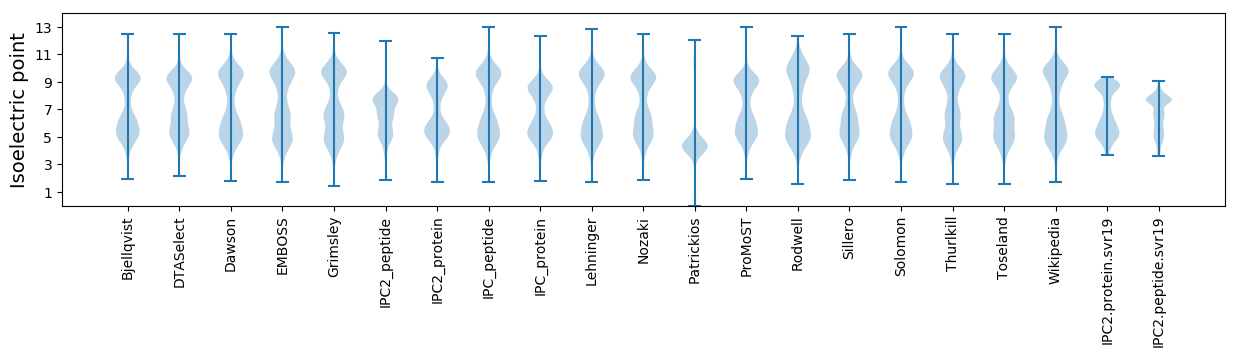
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4W4B9|A0A1M4W4B9_9FLAO Nucleotide-binding universal stress protein UspA family OS=Psychroflexus salarius OX=1155689 GN=SAMN05444278_10575 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.09RR21 pKa = 11.84MASKK25 pKa = 10.6NGRR28 pKa = 11.84KK29 pKa = 9.11VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.67GRR39 pKa = 11.84KK40 pKa = 7.79KK41 pKa = 10.7LSVSSEE47 pKa = 2.78ISMRR51 pKa = 11.84KK52 pKa = 8.72RR53 pKa = 3.06
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.09RR21 pKa = 11.84MASKK25 pKa = 10.6NGRR28 pKa = 11.84KK29 pKa = 9.11VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.67GRR39 pKa = 11.84KK40 pKa = 7.79KK41 pKa = 10.7LSVSSEE47 pKa = 2.78ISMRR51 pKa = 11.84KK52 pKa = 8.72RR53 pKa = 3.06
Molecular weight: 6.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
777795 |
32 |
2412 |
336.6 |
38.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.334 ± 0.05 | 0.708 ± 0.016 |
5.564 ± 0.045 | 6.496 ± 0.044 |
5.379 ± 0.04 | 5.844 ± 0.056 |
1.869 ± 0.024 | 7.858 ± 0.047 |
7.772 ± 0.081 | 9.675 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.948 ± 0.027 | 6.472 ± 0.06 |
3.264 ± 0.025 | 4.235 ± 0.038 |
3.281 ± 0.034 | 6.815 ± 0.043 |
5.598 ± 0.055 | 5.99 ± 0.044 |
0.949 ± 0.019 | 3.947 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
