
Winogradskyella sp. J14-2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella; unclassified Winogradskyella
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
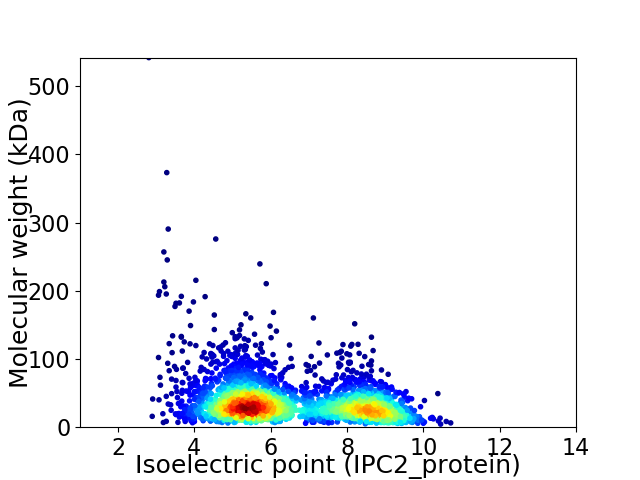
Virtual 2D-PAGE plot for 2928 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
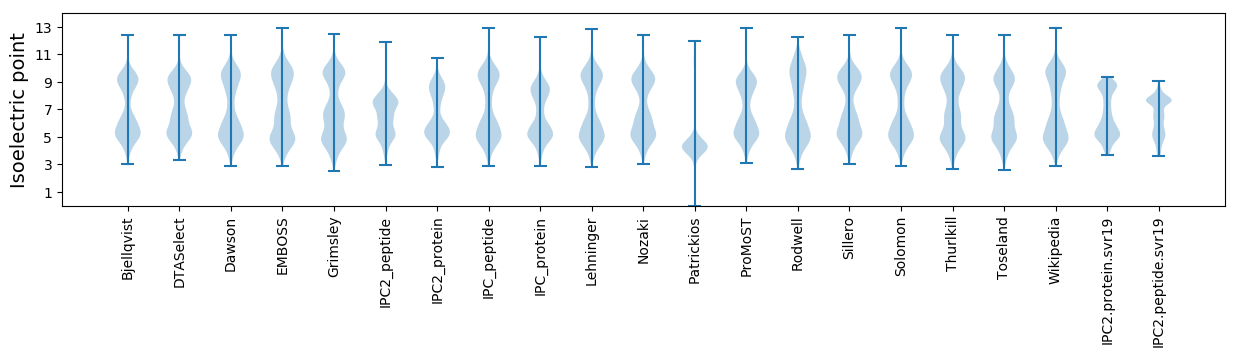
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
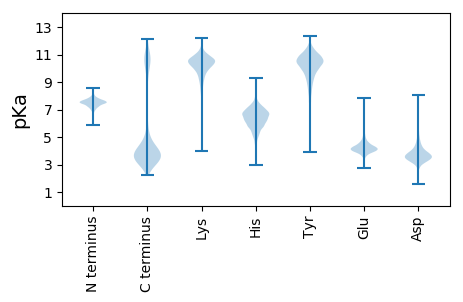
>tr|A0A1P8RU77|A0A1P8RU77_9FLAO Chaperone protein ClpB OS=Winogradskyella sp. J14-2 OX=1936080 GN=clpB PE=3 SV=1
MM1 pKa = 7.57KK2 pKa = 10.14SIKK5 pKa = 10.3LILSLAVVLTLTISCEE21 pKa = 4.15DD22 pKa = 4.31DD23 pKa = 4.03DD24 pKa = 4.84VAVPDD29 pKa = 3.59VEE31 pKa = 4.2VRR33 pKa = 11.84DD34 pKa = 3.88RR35 pKa = 11.84GEE37 pKa = 3.61QQIADD42 pKa = 3.82NDD44 pKa = 4.07SLLGYY49 pKa = 10.61LSTHH53 pKa = 6.6YY54 pKa = 10.73YY55 pKa = 10.66NSDD58 pKa = 3.36FFLTGTNHH66 pKa = 7.35KK67 pKa = 8.99YY68 pKa = 8.92TDD70 pKa = 3.31IVVTEE75 pKa = 4.07LVEE78 pKa = 4.53GEE80 pKa = 4.52VVPAGHH86 pKa = 6.72TLLINAVEE94 pKa = 4.22THH96 pKa = 4.94TTTFEE101 pKa = 4.01EE102 pKa = 3.8TDD104 pKa = 3.34YY105 pKa = 11.32QYY107 pKa = 11.92YY108 pKa = 9.83VLKK111 pKa = 10.51LNQGAGDD118 pKa = 3.86SPRR121 pKa = 11.84FTDD124 pKa = 4.4FVRR127 pKa = 11.84MRR129 pKa = 11.84YY130 pKa = 9.59EE131 pKa = 4.17GFTLEE136 pKa = 4.61DD137 pKa = 3.72GEE139 pKa = 4.83VFDD142 pKa = 6.13SRR144 pKa = 11.84ATPEE148 pKa = 4.42DD149 pKa = 4.07LPMQGVGFSGGVIRR163 pKa = 11.84GWQLTIPMFNTAEE176 pKa = 4.09SFAIGSDD183 pKa = 4.21GITNYY188 pKa = 10.83NNFGLGMMFLPSGLAYY204 pKa = 10.52FSNTASGSVAAYY216 pKa = 10.2SNLVFKK222 pKa = 11.02FEE224 pKa = 4.16LLQYY228 pKa = 10.75EE229 pKa = 4.79EE230 pKa = 4.99IDD232 pKa = 3.54HH233 pKa = 7.27DD234 pKa = 4.62GDD236 pKa = 4.79GIPSYY241 pKa = 11.02IEE243 pKa = 5.22DD244 pKa = 4.17LDD246 pKa = 4.82ANLDD250 pKa = 3.86VLDD253 pKa = 5.78DD254 pKa = 4.42DD255 pKa = 4.55TDD257 pKa = 3.56EE258 pKa = 5.45DD259 pKa = 4.26LAPNYY264 pKa = 10.52VDD266 pKa = 5.36VDD268 pKa = 4.06DD269 pKa = 6.48DD270 pKa = 4.36NDD272 pKa = 3.98GVNTRR277 pKa = 11.84DD278 pKa = 3.23EE279 pKa = 4.59LMPTSYY285 pKa = 10.24TVDD288 pKa = 2.82TNMNEE293 pKa = 4.13EE294 pKa = 4.29EE295 pKa = 4.27PVLGPGEE302 pKa = 4.14YY303 pKa = 9.89EE304 pKa = 3.84ISRR307 pKa = 11.84STSNGILTINTVKK320 pKa = 10.65VVDD323 pKa = 4.05SDD325 pKa = 4.76SNGTPDD331 pKa = 4.4YY332 pKa = 10.08LQEE335 pKa = 5.77DD336 pKa = 3.28IAEE339 pKa = 4.43NYY341 pKa = 10.04NEE343 pKa = 4.17EE344 pKa = 4.24EE345 pKa = 4.4GG346 pKa = 3.89
MM1 pKa = 7.57KK2 pKa = 10.14SIKK5 pKa = 10.3LILSLAVVLTLTISCEE21 pKa = 4.15DD22 pKa = 4.31DD23 pKa = 4.03DD24 pKa = 4.84VAVPDD29 pKa = 3.59VEE31 pKa = 4.2VRR33 pKa = 11.84DD34 pKa = 3.88RR35 pKa = 11.84GEE37 pKa = 3.61QQIADD42 pKa = 3.82NDD44 pKa = 4.07SLLGYY49 pKa = 10.61LSTHH53 pKa = 6.6YY54 pKa = 10.73YY55 pKa = 10.66NSDD58 pKa = 3.36FFLTGTNHH66 pKa = 7.35KK67 pKa = 8.99YY68 pKa = 8.92TDD70 pKa = 3.31IVVTEE75 pKa = 4.07LVEE78 pKa = 4.53GEE80 pKa = 4.52VVPAGHH86 pKa = 6.72TLLINAVEE94 pKa = 4.22THH96 pKa = 4.94TTTFEE101 pKa = 4.01EE102 pKa = 3.8TDD104 pKa = 3.34YY105 pKa = 11.32QYY107 pKa = 11.92YY108 pKa = 9.83VLKK111 pKa = 10.51LNQGAGDD118 pKa = 3.86SPRR121 pKa = 11.84FTDD124 pKa = 4.4FVRR127 pKa = 11.84MRR129 pKa = 11.84YY130 pKa = 9.59EE131 pKa = 4.17GFTLEE136 pKa = 4.61DD137 pKa = 3.72GEE139 pKa = 4.83VFDD142 pKa = 6.13SRR144 pKa = 11.84ATPEE148 pKa = 4.42DD149 pKa = 4.07LPMQGVGFSGGVIRR163 pKa = 11.84GWQLTIPMFNTAEE176 pKa = 4.09SFAIGSDD183 pKa = 4.21GITNYY188 pKa = 10.83NNFGLGMMFLPSGLAYY204 pKa = 10.52FSNTASGSVAAYY216 pKa = 10.2SNLVFKK222 pKa = 11.02FEE224 pKa = 4.16LLQYY228 pKa = 10.75EE229 pKa = 4.79EE230 pKa = 4.99IDD232 pKa = 3.54HH233 pKa = 7.27DD234 pKa = 4.62GDD236 pKa = 4.79GIPSYY241 pKa = 11.02IEE243 pKa = 5.22DD244 pKa = 4.17LDD246 pKa = 4.82ANLDD250 pKa = 3.86VLDD253 pKa = 5.78DD254 pKa = 4.42DD255 pKa = 4.55TDD257 pKa = 3.56EE258 pKa = 5.45DD259 pKa = 4.26LAPNYY264 pKa = 10.52VDD266 pKa = 5.36VDD268 pKa = 4.06DD269 pKa = 6.48DD270 pKa = 4.36NDD272 pKa = 3.98GVNTRR277 pKa = 11.84DD278 pKa = 3.23EE279 pKa = 4.59LMPTSYY285 pKa = 10.24TVDD288 pKa = 2.82TNMNEE293 pKa = 4.13EE294 pKa = 4.29EE295 pKa = 4.27PVLGPGEE302 pKa = 4.14YY303 pKa = 9.89EE304 pKa = 3.84ISRR307 pKa = 11.84STSNGILTINTVKK320 pKa = 10.65VVDD323 pKa = 4.05SDD325 pKa = 4.76SNGTPDD331 pKa = 4.4YY332 pKa = 10.08LQEE335 pKa = 5.77DD336 pKa = 3.28IAEE339 pKa = 4.43NYY341 pKa = 10.04NEE343 pKa = 4.17EE344 pKa = 4.24EE345 pKa = 4.4GG346 pKa = 3.89
Molecular weight: 38.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8RZT9|A0A1P8RZT9_9FLAO GTP cyclohydrolase 1 OS=Winogradskyella sp. J14-2 OX=1936080 GN=folE PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.18VLASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.66LSVSSEE48 pKa = 3.9LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.18RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.18VLASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.66LSVSSEE48 pKa = 3.9LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1008313 |
38 |
5318 |
344.4 |
38.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.311 ± 0.048 | 0.769 ± 0.02 |
6.002 ± 0.05 | 6.534 ± 0.044 |
5.218 ± 0.038 | 6.317 ± 0.057 |
1.706 ± 0.028 | 7.934 ± 0.048 |
7.594 ± 0.09 | 9.118 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.038 ± 0.029 | 6.622 ± 0.055 |
3.299 ± 0.024 | 3.26 ± 0.026 |
3.339 ± 0.037 | 6.425 ± 0.043 |
6.108 ± 0.089 | 6.238 ± 0.035 |
1.016 ± 0.017 | 4.151 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
