
Fluviicola taffensis (strain DSM 16823 / NCIMB 13979 / RW262)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Crocinitomicaceae; Fluviicola; Fluviicola taffensis
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
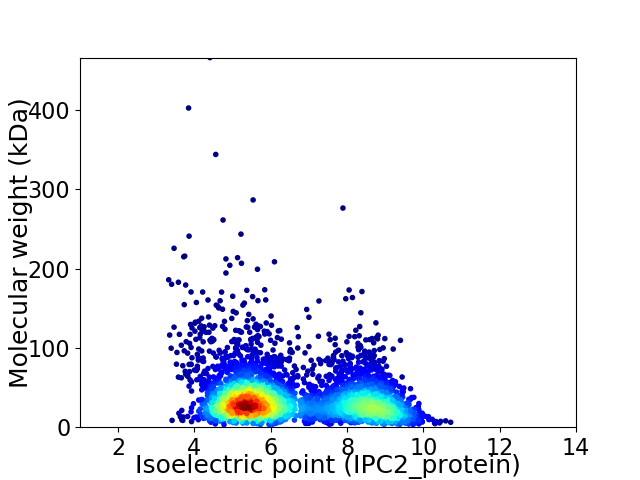
Virtual 2D-PAGE plot for 4016 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2IEW8|F2IEW8_FLUTR Uncharacterized protein OS=Fluviicola taffensis (strain DSM 16823 / NCIMB 13979 / RW262) OX=755732 GN=Fluta_0426 PE=4 SV=1
MM1 pKa = 7.75KK2 pKa = 10.59SPLQWLFVASITMVSGISFSQTRR25 pKa = 11.84YY26 pKa = 10.04VSTTGTDD33 pKa = 3.12VSNDD37 pKa = 3.27CQTPGNPCGTISHH50 pKa = 7.33AYY52 pKa = 9.73SEE54 pKa = 5.48AIADD58 pKa = 4.07DD59 pKa = 4.42TIKK62 pKa = 10.47IAPGNYY68 pKa = 9.62NLTSTLSLLQPVVIAALDD86 pKa = 3.9PMDD89 pKa = 5.0LPVITSSASDD99 pKa = 3.8VISVNSDD106 pKa = 2.96DD107 pKa = 3.71VTIMNLHH114 pKa = 6.62FEE116 pKa = 4.35MGLTASTGLRR126 pKa = 11.84GIVANTGDD134 pKa = 3.98FDD136 pKa = 3.99NLKK139 pKa = 10.07VSGNIFEE146 pKa = 4.59STNPISGTPNTNMVWTAFAISLVATSGDD174 pKa = 3.67VQNVTIEE181 pKa = 4.17DD182 pKa = 3.69NEE184 pKa = 4.42IIADD188 pKa = 3.53ATSNVFGRR196 pKa = 11.84GIFLGNGGTGLDD208 pKa = 3.85APGGTISGNTIKK220 pKa = 10.36AYY222 pKa = 10.51YY223 pKa = 9.57AIQSALIADD232 pKa = 4.68DD233 pKa = 5.87LSISDD238 pKa = 4.08NDD240 pKa = 3.18IDD242 pKa = 4.1GTVLIIAPKK251 pKa = 10.04AGVIVDD257 pKa = 4.12VEE259 pKa = 4.59DD260 pKa = 4.17NMLGNNTQMTPASYY274 pKa = 10.69YY275 pKa = 10.79AVLEE279 pKa = 4.06LRR281 pKa = 11.84SIEE284 pKa = 4.11VGDD287 pKa = 3.8VNVSGNQFSNYY298 pKa = 8.56VNIGLLSEE306 pKa = 4.08ASKK309 pKa = 11.02NVSVLGNIFTPDD321 pKa = 2.96AAASGFTSIMANTKK335 pKa = 10.6LMTNGAQGNTYY346 pKa = 10.26SDD348 pKa = 4.59EE349 pKa = 3.87ITIQGNTFNAGLTGQGTAILFADD372 pKa = 4.22HH373 pKa = 6.75YY374 pKa = 11.08GVNTPAFGNITIGGAGVTDD393 pKa = 3.59KK394 pKa = 10.89NTFNSNLGTYY404 pKa = 9.41IALDD408 pKa = 3.92EE409 pKa = 4.53LSGPSTAFPLWATYY423 pKa = 8.52PATTMAPFSQDD434 pKa = 2.69VEE436 pKa = 4.35AYY438 pKa = 10.17AIYY441 pKa = 10.94NNYY444 pKa = 9.74GLANFAAIEE453 pKa = 4.26AKK455 pKa = 10.93NNDD458 pKa = 3.41QSDD461 pKa = 3.64EE462 pKa = 4.21AGLGKK467 pKa = 9.98IYY469 pKa = 9.71LTAYY473 pKa = 10.0GEE475 pKa = 4.0NRR477 pKa = 11.84YY478 pKa = 10.03VSTTGTDD485 pKa = 3.48VLNTCTLPGSPCLTIAHH502 pKa = 7.59AISVAIDD509 pKa = 3.45EE510 pKa = 5.2DD511 pKa = 4.43SVIVYY516 pKa = 8.99PGSYY520 pKa = 10.94ALTSVLNIGQDD531 pKa = 3.79GLVLTASDD539 pKa = 4.24VNDD542 pKa = 3.75KK543 pKa = 10.87PVITTNQANIIDD555 pKa = 4.12VNAEE559 pKa = 3.9DD560 pKa = 3.65VTVNGFEE567 pKa = 4.32FRR569 pKa = 11.84MGLTNATGIKK579 pKa = 10.1GVVSTASNFNNLKK592 pKa = 9.8VSKK595 pKa = 10.75NDD597 pKa = 4.34FISTAALDD605 pKa = 3.92LEE607 pKa = 4.42GMVWSAFAVQLTSATGEE624 pKa = 3.85IDD626 pKa = 3.34TVEE629 pKa = 3.92ITDD632 pKa = 4.08NGITTAATNLNVFGRR647 pKa = 11.84GFYY650 pKa = 10.47LGSGSANGPGGLIANNQIQAYY671 pKa = 9.03YY672 pKa = 10.5AIQTANIATDD682 pKa = 3.64LTVEE686 pKa = 4.27SNDD689 pKa = 3.12INGISMFSAPLNNSLFDD706 pKa = 3.76VYY708 pKa = 11.72DD709 pKa = 3.58NTFSGANMNTNMTSDD724 pKa = 2.95SLYY727 pKa = 10.56AVVEE731 pKa = 3.85IRR733 pKa = 11.84GIASGSIEE741 pKa = 4.81FYY743 pKa = 11.16DD744 pKa = 3.55NTIEE748 pKa = 4.22GFSKK752 pKa = 10.36IGLLSSASKK761 pKa = 10.88DD762 pKa = 3.65VIVDD766 pKa = 3.62ANVFTPKK773 pKa = 9.52ATATAFTSLMANTKK787 pKa = 10.57LMTNGVQGNTYY798 pKa = 10.1SDD800 pKa = 4.48EE801 pKa = 3.78ITIIGNTFSAGATNGGTAIIFADD824 pKa = 3.72HH825 pKa = 6.55YY826 pKa = 11.31GVNTPAFADD835 pKa = 3.48ITVGGAATAEE845 pKa = 4.12KK846 pKa = 10.81NIFNASLGKK855 pKa = 10.81YY856 pKa = 9.04IVLDD860 pKa = 3.7NKK862 pKa = 10.14TGTSASEE869 pKa = 4.03PLWAVYY875 pKa = 10.22DD876 pKa = 3.86ATNMKK881 pKa = 9.98PVTQNVEE888 pKa = 3.57ALYY891 pKa = 11.33VNNTYY896 pKa = 11.2NFGSTGEE903 pKa = 4.21VEE905 pKa = 4.71LKK907 pKa = 11.02NIDD910 pKa = 3.64SVDD913 pKa = 3.42NVILGKK919 pKa = 10.85VILAYY924 pKa = 10.7SFVGLDD930 pKa = 3.52EE931 pKa = 4.52LTSADD936 pKa = 3.06ATLYY940 pKa = 9.53PNPAINAVTIEE951 pKa = 4.22LTDD954 pKa = 3.99LNAVVEE960 pKa = 4.38MTVVDD965 pKa = 3.79LVGNVVYY972 pKa = 10.92NSTINGSITVDD983 pKa = 3.34VSNLTSGVYY992 pKa = 8.58VVRR995 pKa = 11.84LNNKK999 pKa = 8.32GQVSSTRR1006 pKa = 11.84FVKK1009 pKa = 10.69NN1010 pKa = 3.09
MM1 pKa = 7.75KK2 pKa = 10.59SPLQWLFVASITMVSGISFSQTRR25 pKa = 11.84YY26 pKa = 10.04VSTTGTDD33 pKa = 3.12VSNDD37 pKa = 3.27CQTPGNPCGTISHH50 pKa = 7.33AYY52 pKa = 9.73SEE54 pKa = 5.48AIADD58 pKa = 4.07DD59 pKa = 4.42TIKK62 pKa = 10.47IAPGNYY68 pKa = 9.62NLTSTLSLLQPVVIAALDD86 pKa = 3.9PMDD89 pKa = 5.0LPVITSSASDD99 pKa = 3.8VISVNSDD106 pKa = 2.96DD107 pKa = 3.71VTIMNLHH114 pKa = 6.62FEE116 pKa = 4.35MGLTASTGLRR126 pKa = 11.84GIVANTGDD134 pKa = 3.98FDD136 pKa = 3.99NLKK139 pKa = 10.07VSGNIFEE146 pKa = 4.59STNPISGTPNTNMVWTAFAISLVATSGDD174 pKa = 3.67VQNVTIEE181 pKa = 4.17DD182 pKa = 3.69NEE184 pKa = 4.42IIADD188 pKa = 3.53ATSNVFGRR196 pKa = 11.84GIFLGNGGTGLDD208 pKa = 3.85APGGTISGNTIKK220 pKa = 10.36AYY222 pKa = 10.51YY223 pKa = 9.57AIQSALIADD232 pKa = 4.68DD233 pKa = 5.87LSISDD238 pKa = 4.08NDD240 pKa = 3.18IDD242 pKa = 4.1GTVLIIAPKK251 pKa = 10.04AGVIVDD257 pKa = 4.12VEE259 pKa = 4.59DD260 pKa = 4.17NMLGNNTQMTPASYY274 pKa = 10.69YY275 pKa = 10.79AVLEE279 pKa = 4.06LRR281 pKa = 11.84SIEE284 pKa = 4.11VGDD287 pKa = 3.8VNVSGNQFSNYY298 pKa = 8.56VNIGLLSEE306 pKa = 4.08ASKK309 pKa = 11.02NVSVLGNIFTPDD321 pKa = 2.96AAASGFTSIMANTKK335 pKa = 10.6LMTNGAQGNTYY346 pKa = 10.26SDD348 pKa = 4.59EE349 pKa = 3.87ITIQGNTFNAGLTGQGTAILFADD372 pKa = 4.22HH373 pKa = 6.75YY374 pKa = 11.08GVNTPAFGNITIGGAGVTDD393 pKa = 3.59KK394 pKa = 10.89NTFNSNLGTYY404 pKa = 9.41IALDD408 pKa = 3.92EE409 pKa = 4.53LSGPSTAFPLWATYY423 pKa = 8.52PATTMAPFSQDD434 pKa = 2.69VEE436 pKa = 4.35AYY438 pKa = 10.17AIYY441 pKa = 10.94NNYY444 pKa = 9.74GLANFAAIEE453 pKa = 4.26AKK455 pKa = 10.93NNDD458 pKa = 3.41QSDD461 pKa = 3.64EE462 pKa = 4.21AGLGKK467 pKa = 9.98IYY469 pKa = 9.71LTAYY473 pKa = 10.0GEE475 pKa = 4.0NRR477 pKa = 11.84YY478 pKa = 10.03VSTTGTDD485 pKa = 3.48VLNTCTLPGSPCLTIAHH502 pKa = 7.59AISVAIDD509 pKa = 3.45EE510 pKa = 5.2DD511 pKa = 4.43SVIVYY516 pKa = 8.99PGSYY520 pKa = 10.94ALTSVLNIGQDD531 pKa = 3.79GLVLTASDD539 pKa = 4.24VNDD542 pKa = 3.75KK543 pKa = 10.87PVITTNQANIIDD555 pKa = 4.12VNAEE559 pKa = 3.9DD560 pKa = 3.65VTVNGFEE567 pKa = 4.32FRR569 pKa = 11.84MGLTNATGIKK579 pKa = 10.1GVVSTASNFNNLKK592 pKa = 9.8VSKK595 pKa = 10.75NDD597 pKa = 4.34FISTAALDD605 pKa = 3.92LEE607 pKa = 4.42GMVWSAFAVQLTSATGEE624 pKa = 3.85IDD626 pKa = 3.34TVEE629 pKa = 3.92ITDD632 pKa = 4.08NGITTAATNLNVFGRR647 pKa = 11.84GFYY650 pKa = 10.47LGSGSANGPGGLIANNQIQAYY671 pKa = 9.03YY672 pKa = 10.5AIQTANIATDD682 pKa = 3.64LTVEE686 pKa = 4.27SNDD689 pKa = 3.12INGISMFSAPLNNSLFDD706 pKa = 3.76VYY708 pKa = 11.72DD709 pKa = 3.58NTFSGANMNTNMTSDD724 pKa = 2.95SLYY727 pKa = 10.56AVVEE731 pKa = 3.85IRR733 pKa = 11.84GIASGSIEE741 pKa = 4.81FYY743 pKa = 11.16DD744 pKa = 3.55NTIEE748 pKa = 4.22GFSKK752 pKa = 10.36IGLLSSASKK761 pKa = 10.88DD762 pKa = 3.65VIVDD766 pKa = 3.62ANVFTPKK773 pKa = 9.52ATATAFTSLMANTKK787 pKa = 10.57LMTNGVQGNTYY798 pKa = 10.1SDD800 pKa = 4.48EE801 pKa = 3.78ITIIGNTFSAGATNGGTAIIFADD824 pKa = 3.72HH825 pKa = 6.55YY826 pKa = 11.31GVNTPAFADD835 pKa = 3.48ITVGGAATAEE845 pKa = 4.12KK846 pKa = 10.81NIFNASLGKK855 pKa = 10.81YY856 pKa = 9.04IVLDD860 pKa = 3.7NKK862 pKa = 10.14TGTSASEE869 pKa = 4.03PLWAVYY875 pKa = 10.22DD876 pKa = 3.86ATNMKK881 pKa = 9.98PVTQNVEE888 pKa = 3.57ALYY891 pKa = 11.33VNNTYY896 pKa = 11.2NFGSTGEE903 pKa = 4.21VEE905 pKa = 4.71LKK907 pKa = 11.02NIDD910 pKa = 3.64SVDD913 pKa = 3.42NVILGKK919 pKa = 10.85VILAYY924 pKa = 10.7SFVGLDD930 pKa = 3.52EE931 pKa = 4.52LTSADD936 pKa = 3.06ATLYY940 pKa = 9.53PNPAINAVTIEE951 pKa = 4.22LTDD954 pKa = 3.99LNAVVEE960 pKa = 4.38MTVVDD965 pKa = 3.79LVGNVVYY972 pKa = 10.92NSTINGSITVDD983 pKa = 3.34VSNLTSGVYY992 pKa = 8.58VVRR995 pKa = 11.84LNNKK999 pKa = 8.32GQVSSTRR1006 pKa = 11.84FVKK1009 pKa = 10.69NN1010 pKa = 3.09
Molecular weight: 105.71 kDa
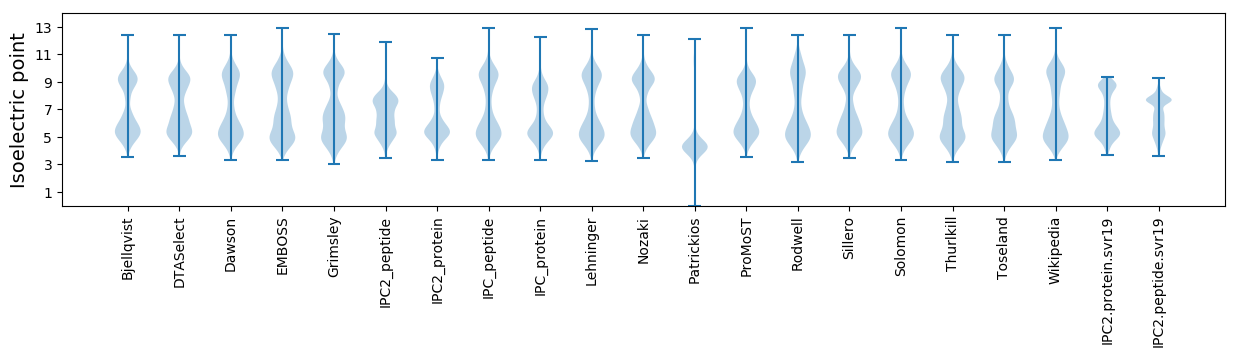
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2IG14|F2IG14_FLUTR Uncharacterized protein OS=Fluviicola taffensis (strain DSM 16823 / NCIMB 13979 / RW262) OX=755732 GN=Fluta_1643 PE=4 SV=1
MM1 pKa = 7.52IFKK4 pKa = 10.76LDD6 pKa = 3.56TTKK9 pKa = 10.83KK10 pKa = 9.77EE11 pKa = 4.13RR12 pKa = 11.84NTATNRR18 pKa = 11.84RR19 pKa = 11.84LVQWRR24 pKa = 11.84VMWLIEE30 pKa = 4.35HH31 pKa = 5.91STSHH35 pKa = 5.72QLLWCIDD42 pKa = 3.68SFVLRR47 pKa = 11.84NPPLRR52 pKa = 11.84QAPNRR57 pKa = 11.84YY58 pKa = 6.67TAALDD63 pKa = 3.93EE64 pKa = 4.21NHH66 pKa = 5.42QTTFRR71 pKa = 11.84QQ72 pKa = 3.44
MM1 pKa = 7.52IFKK4 pKa = 10.76LDD6 pKa = 3.56TTKK9 pKa = 10.83KK10 pKa = 9.77EE11 pKa = 4.13RR12 pKa = 11.84NTATNRR18 pKa = 11.84RR19 pKa = 11.84LVQWRR24 pKa = 11.84VMWLIEE30 pKa = 4.35HH31 pKa = 5.91STSHH35 pKa = 5.72QLLWCIDD42 pKa = 3.68SFVLRR47 pKa = 11.84NPPLRR52 pKa = 11.84QAPNRR57 pKa = 11.84YY58 pKa = 6.67TAALDD63 pKa = 3.93EE64 pKa = 4.21NHH66 pKa = 5.42QTTFRR71 pKa = 11.84QQ72 pKa = 3.44
Molecular weight: 8.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1385128 |
30 |
4221 |
344.9 |
38.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.14 ± 0.044 | 1.005 ± 0.017 |
5.159 ± 0.028 | 6.187 ± 0.046 |
5.302 ± 0.036 | 6.681 ± 0.04 |
1.779 ± 0.019 | 7.72 ± 0.031 |
6.99 ± 0.055 | 9.24 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.197 ± 0.02 | 5.975 ± 0.041 |
3.56 ± 0.027 | 3.723 ± 0.022 |
3.481 ± 0.029 | 7.203 ± 0.039 |
6.289 ± 0.071 | 6.219 ± 0.033 |
1.153 ± 0.015 | 3.997 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
