
Caenorhabditis latens
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Peloderinae; Caenorhabditis
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
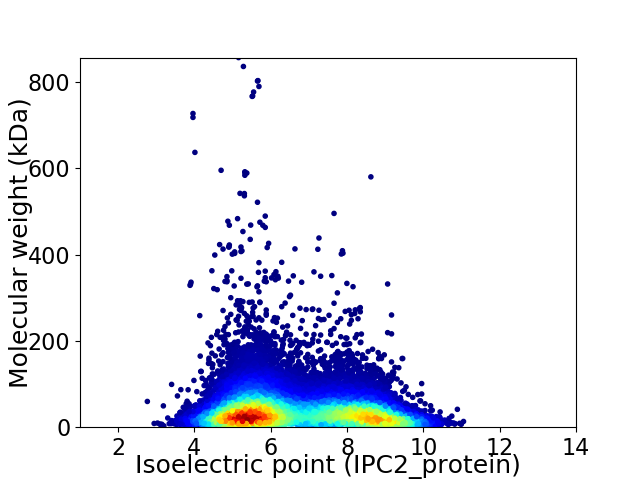
Virtual 2D-PAGE plot for 25394 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A261BFL7|A0A261BFL7_9PELO Uncharacterized protein (Fragment) OS=Caenorhabditis latens OX=1503980 GN=FL83_16552 PE=4 SV=1
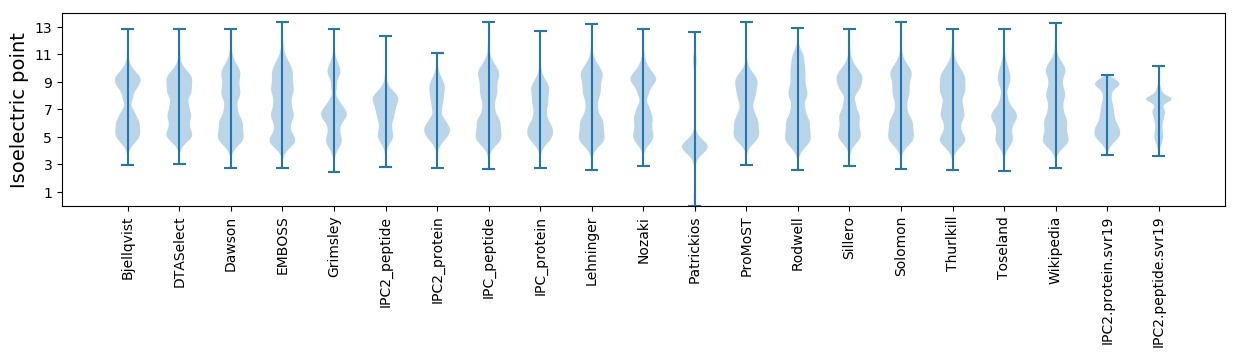
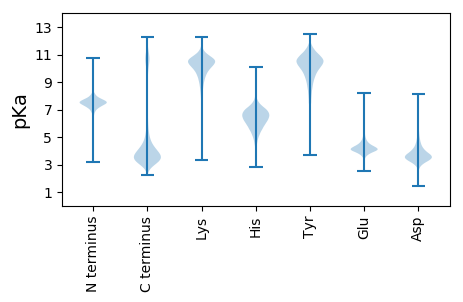
MM1 pKa = 6.96NQNTYY6 pKa = 10.71SADD9 pKa = 3.51SRR11 pKa = 11.84GGTINYY17 pKa = 7.58TVTNICNFVVIPHH30 pKa = 6.4SFAAPPFCPPSGNPPTSNYY49 pKa = 8.81HH50 pKa = 4.67QNEE53 pKa = 4.09GASRR57 pKa = 11.84SRR59 pKa = 11.84GDD61 pKa = 3.45GEE63 pKa = 4.79NDD65 pKa = 3.32YY66 pKa = 11.71GDD68 pKa = 4.4AGAPNTPSNDD78 pKa = 3.23NTDD81 pKa = 4.07PEE83 pKa = 4.36GHH85 pKa = 6.72PVDD88 pKa = 5.26GIRR91 pKa = 11.84ITGMPTALIALPYY104 pKa = 9.2LDD106 pKa = 4.95PMVDD110 pKa = 3.29STRR113 pKa = 11.84LVAIAAEE120 pKa = 4.2NRR122 pKa = 11.84SPLTTDD128 pKa = 4.58ALITSNNPSSVATHH142 pKa = 5.92TEE144 pKa = 3.71VSDD147 pKa = 3.66AEE149 pKa = 4.24RR150 pKa = 11.84NIAEE154 pKa = 4.22YY155 pKa = 9.85TGEE158 pKa = 4.04SDD160 pKa = 4.43EE161 pKa = 4.79EE162 pKa = 4.6PNTAEE167 pKa = 4.0PAGTHH172 pKa = 3.89STEE175 pKa = 4.19YY176 pKa = 8.98EE177 pKa = 3.91TTNAVTAEE185 pKa = 4.11TANLEE190 pKa = 4.24RR191 pKa = 11.84SDD193 pKa = 3.41TAAAAEE199 pKa = 4.27SGVIQTVDD207 pKa = 3.97FEE209 pKa = 4.63SADD212 pKa = 3.47AEE214 pKa = 4.64TYY216 pKa = 10.69SVAEE220 pKa = 4.18TAEE223 pKa = 4.47TPTSDD228 pKa = 5.13AEE230 pKa = 4.37TADD233 pKa = 3.69TQLVDD238 pKa = 3.88VRR240 pKa = 11.84MDD242 pKa = 4.03DD243 pKa = 5.55DD244 pKa = 3.8EE245 pKa = 4.47TADD248 pKa = 3.82AVIAGYY254 pKa = 10.18QLTDD258 pKa = 4.63DD259 pKa = 5.13PDD261 pKa = 6.04ADD263 pKa = 4.32TPNADD268 pKa = 3.6PQVTSVQMNHH278 pKa = 6.14TPDD281 pKa = 3.69ADD283 pKa = 3.53AGLIDD288 pKa = 4.2FQLDD292 pKa = 3.66DD293 pKa = 3.62AHH295 pKa = 6.62MEE297 pKa = 4.03QNLDD301 pKa = 3.49VVADD305 pKa = 3.99GLEE308 pKa = 4.21TEE310 pKa = 4.66SAISGALLFNN320 pKa = 4.92
MM1 pKa = 6.96NQNTYY6 pKa = 10.71SADD9 pKa = 3.51SRR11 pKa = 11.84GGTINYY17 pKa = 7.58TVTNICNFVVIPHH30 pKa = 6.4SFAAPPFCPPSGNPPTSNYY49 pKa = 8.81HH50 pKa = 4.67QNEE53 pKa = 4.09GASRR57 pKa = 11.84SRR59 pKa = 11.84GDD61 pKa = 3.45GEE63 pKa = 4.79NDD65 pKa = 3.32YY66 pKa = 11.71GDD68 pKa = 4.4AGAPNTPSNDD78 pKa = 3.23NTDD81 pKa = 4.07PEE83 pKa = 4.36GHH85 pKa = 6.72PVDD88 pKa = 5.26GIRR91 pKa = 11.84ITGMPTALIALPYY104 pKa = 9.2LDD106 pKa = 4.95PMVDD110 pKa = 3.29STRR113 pKa = 11.84LVAIAAEE120 pKa = 4.2NRR122 pKa = 11.84SPLTTDD128 pKa = 4.58ALITSNNPSSVATHH142 pKa = 5.92TEE144 pKa = 3.71VSDD147 pKa = 3.66AEE149 pKa = 4.24RR150 pKa = 11.84NIAEE154 pKa = 4.22YY155 pKa = 9.85TGEE158 pKa = 4.04SDD160 pKa = 4.43EE161 pKa = 4.79EE162 pKa = 4.6PNTAEE167 pKa = 4.0PAGTHH172 pKa = 3.89STEE175 pKa = 4.19YY176 pKa = 8.98EE177 pKa = 3.91TTNAVTAEE185 pKa = 4.11TANLEE190 pKa = 4.24RR191 pKa = 11.84SDD193 pKa = 3.41TAAAAEE199 pKa = 4.27SGVIQTVDD207 pKa = 3.97FEE209 pKa = 4.63SADD212 pKa = 3.47AEE214 pKa = 4.64TYY216 pKa = 10.69SVAEE220 pKa = 4.18TAEE223 pKa = 4.47TPTSDD228 pKa = 5.13AEE230 pKa = 4.37TADD233 pKa = 3.69TQLVDD238 pKa = 3.88VRR240 pKa = 11.84MDD242 pKa = 4.03DD243 pKa = 5.55DD244 pKa = 3.8EE245 pKa = 4.47TADD248 pKa = 3.82AVIAGYY254 pKa = 10.18QLTDD258 pKa = 4.63DD259 pKa = 5.13PDD261 pKa = 6.04ADD263 pKa = 4.32TPNADD268 pKa = 3.6PQVTSVQMNHH278 pKa = 6.14TPDD281 pKa = 3.69ADD283 pKa = 3.53AGLIDD288 pKa = 4.2FQLDD292 pKa = 3.66DD293 pKa = 3.62AHH295 pKa = 6.62MEE297 pKa = 4.03QNLDD301 pKa = 3.49VVADD305 pKa = 3.99GLEE308 pKa = 4.21TEE310 pKa = 4.66SAISGALLFNN320 pKa = 4.92
Molecular weight: 33.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A261CEL6|A0A261CEL6_9PELO Protein kinase domain-containing protein (Fragment) OS=Caenorhabditis latens OX=1503980 GN=FL83_04616 PE=4 SV=1
MM1 pKa = 7.24TRR3 pKa = 11.84GTAPRR8 pKa = 11.84HH9 pKa = 5.14RR10 pKa = 11.84PPRR13 pKa = 11.84PATPRR18 pKa = 11.84QPPAAIVHH26 pKa = 6.45HH27 pKa = 6.39SPPNNSTFSRR37 pKa = 11.84HH38 pKa = 4.01RR39 pKa = 11.84SVRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84IQMGRR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84TSPRR56 pKa = 11.84RR57 pKa = 11.84PDD59 pKa = 3.22SPRR62 pKa = 11.84TKK64 pKa = 10.61LL65 pKa = 3.37
MM1 pKa = 7.24TRR3 pKa = 11.84GTAPRR8 pKa = 11.84HH9 pKa = 5.14RR10 pKa = 11.84PPRR13 pKa = 11.84PATPRR18 pKa = 11.84QPPAAIVHH26 pKa = 6.45HH27 pKa = 6.39SPPNNSTFSRR37 pKa = 11.84HH38 pKa = 4.01RR39 pKa = 11.84SVRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84IQMGRR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84TSPRR56 pKa = 11.84RR57 pKa = 11.84PDD59 pKa = 3.22SPRR62 pKa = 11.84TKK64 pKa = 10.61LL65 pKa = 3.37
Molecular weight: 7.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9829288 |
17 |
7710 |
387.1 |
43.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.056 ± 0.018 | 1.946 ± 0.013 |
5.421 ± 0.015 | 7.061 ± 0.026 |
4.535 ± 0.017 | 5.266 ± 0.019 |
2.316 ± 0.009 | 5.985 ± 0.015 |
6.508 ± 0.021 | 8.469 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.652 ± 0.007 | 4.887 ± 0.011 |
5.014 ± 0.019 | 4.074 ± 0.016 |
5.359 ± 0.016 | 7.979 ± 0.02 |
5.956 ± 0.021 | 6.254 ± 0.016 |
1.112 ± 0.005 | 3.121 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
