
Cyclovirus Chimp11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Chimpanzee associated cyclovirus 1
Average proteome isoelectric point is 8.74
Get precalculated fractions of proteins
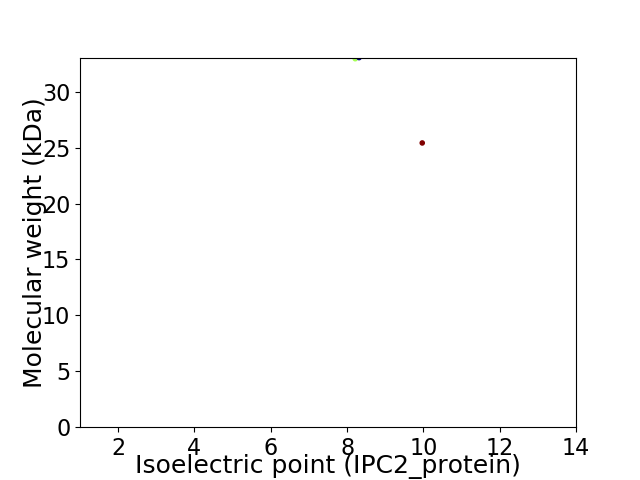
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4N3P7|D4N3P7_9CIRC Capsid protein OS=Cyclovirus Chimp11 OX=742920 GN=cap PE=4 SV=1
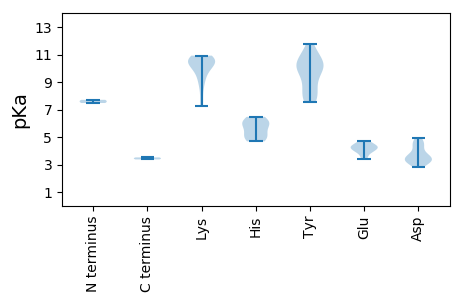
MM1 pKa = 7.71SNRR4 pKa = 11.84TVRR7 pKa = 11.84KK8 pKa = 9.49FCFTWNNYY16 pKa = 8.61EE17 pKa = 3.44FDD19 pKa = 4.46AYY21 pKa = 10.49AKK23 pKa = 10.9CEE25 pKa = 3.93TFLNNFAKK33 pKa = 10.57YY34 pKa = 10.03GIVGEE39 pKa = 4.34EE40 pKa = 4.3LCPSTGTPHH49 pKa = 6.18LQGYY53 pKa = 9.24VNLIKK58 pKa = 9.37PTRR61 pKa = 11.84FSTIKK66 pKa = 10.19KK67 pKa = 8.11HH68 pKa = 5.11LHH70 pKa = 4.87NAIHH74 pKa = 6.47IEE76 pKa = 3.94KK77 pKa = 10.73ANGSDD82 pKa = 3.6EE83 pKa = 4.48QNQTYY88 pKa = 9.65CRR90 pKa = 11.84KK91 pKa = 10.23SGIFFEE97 pKa = 4.68KK98 pKa = 10.77GEE100 pKa = 4.58PIKK103 pKa = 10.55QGQRR107 pKa = 11.84TDD109 pKa = 3.27LQLLVKK115 pKa = 10.11DD116 pKa = 3.73TMEE119 pKa = 4.65PSNTLKK125 pKa = 10.83DD126 pKa = 3.28IATKK130 pKa = 10.65HH131 pKa = 5.76PIAYY135 pKa = 8.54IRR137 pKa = 11.84YY138 pKa = 7.9FRR140 pKa = 11.84GIQEE144 pKa = 4.4LRR146 pKa = 11.84RR147 pKa = 11.84MVLPVPPRR155 pKa = 11.84NYY157 pKa = 7.83PTEE160 pKa = 3.67VRR162 pKa = 11.84YY163 pKa = 10.06YY164 pKa = 9.21WGPPGSGKK172 pKa = 8.76SRR174 pKa = 11.84RR175 pKa = 11.84ALQEE179 pKa = 3.42ATEE182 pKa = 4.05ISTNGIYY189 pKa = 10.21YY190 pKa = 10.12KK191 pKa = 10.53PRR193 pKa = 11.84GQWWDD198 pKa = 3.42GYY200 pKa = 7.55EE201 pKa = 3.96QQSCVIIDD209 pKa = 4.9DD210 pKa = 4.45FYY212 pKa = 11.76GWIKK216 pKa = 10.51YY217 pKa = 10.15DD218 pKa = 4.69EE219 pKa = 4.23ILKK222 pKa = 10.17ICDD225 pKa = 3.22RR226 pKa = 11.84YY227 pKa = 9.85PYY229 pKa = 10.1KK230 pKa = 10.91VQIKK234 pKa = 10.07GGYY237 pKa = 8.28EE238 pKa = 4.0EE239 pKa = 4.65FTSTHH244 pKa = 4.68IWFTSNVDD252 pKa = 2.84TDD254 pKa = 4.26LLYY257 pKa = 11.15KK258 pKa = 10.08FNNYY262 pKa = 9.13INTAFEE268 pKa = 3.72RR269 pKa = 11.84RR270 pKa = 11.84ITIKK274 pKa = 10.45EE275 pKa = 4.27HH276 pKa = 5.24ITLSS280 pKa = 3.52
MM1 pKa = 7.71SNRR4 pKa = 11.84TVRR7 pKa = 11.84KK8 pKa = 9.49FCFTWNNYY16 pKa = 8.61EE17 pKa = 3.44FDD19 pKa = 4.46AYY21 pKa = 10.49AKK23 pKa = 10.9CEE25 pKa = 3.93TFLNNFAKK33 pKa = 10.57YY34 pKa = 10.03GIVGEE39 pKa = 4.34EE40 pKa = 4.3LCPSTGTPHH49 pKa = 6.18LQGYY53 pKa = 9.24VNLIKK58 pKa = 9.37PTRR61 pKa = 11.84FSTIKK66 pKa = 10.19KK67 pKa = 8.11HH68 pKa = 5.11LHH70 pKa = 4.87NAIHH74 pKa = 6.47IEE76 pKa = 3.94KK77 pKa = 10.73ANGSDD82 pKa = 3.6EE83 pKa = 4.48QNQTYY88 pKa = 9.65CRR90 pKa = 11.84KK91 pKa = 10.23SGIFFEE97 pKa = 4.68KK98 pKa = 10.77GEE100 pKa = 4.58PIKK103 pKa = 10.55QGQRR107 pKa = 11.84TDD109 pKa = 3.27LQLLVKK115 pKa = 10.11DD116 pKa = 3.73TMEE119 pKa = 4.65PSNTLKK125 pKa = 10.83DD126 pKa = 3.28IATKK130 pKa = 10.65HH131 pKa = 5.76PIAYY135 pKa = 8.54IRR137 pKa = 11.84YY138 pKa = 7.9FRR140 pKa = 11.84GIQEE144 pKa = 4.4LRR146 pKa = 11.84RR147 pKa = 11.84MVLPVPPRR155 pKa = 11.84NYY157 pKa = 7.83PTEE160 pKa = 3.67VRR162 pKa = 11.84YY163 pKa = 10.06YY164 pKa = 9.21WGPPGSGKK172 pKa = 8.76SRR174 pKa = 11.84RR175 pKa = 11.84ALQEE179 pKa = 3.42ATEE182 pKa = 4.05ISTNGIYY189 pKa = 10.21YY190 pKa = 10.12KK191 pKa = 10.53PRR193 pKa = 11.84GQWWDD198 pKa = 3.42GYY200 pKa = 7.55EE201 pKa = 3.96QQSCVIIDD209 pKa = 4.9DD210 pKa = 4.45FYY212 pKa = 11.76GWIKK216 pKa = 10.51YY217 pKa = 10.15DD218 pKa = 4.69EE219 pKa = 4.23ILKK222 pKa = 10.17ICDD225 pKa = 3.22RR226 pKa = 11.84YY227 pKa = 9.85PYY229 pKa = 10.1KK230 pKa = 10.91VQIKK234 pKa = 10.07GGYY237 pKa = 8.28EE238 pKa = 4.0EE239 pKa = 4.65FTSTHH244 pKa = 4.68IWFTSNVDD252 pKa = 2.84TDD254 pKa = 4.26LLYY257 pKa = 11.15KK258 pKa = 10.08FNNYY262 pKa = 9.13INTAFEE268 pKa = 3.72RR269 pKa = 11.84RR270 pKa = 11.84ITIKK274 pKa = 10.45EE275 pKa = 4.27HH276 pKa = 5.24ITLSS280 pKa = 3.52
Molecular weight: 32.95 kDa
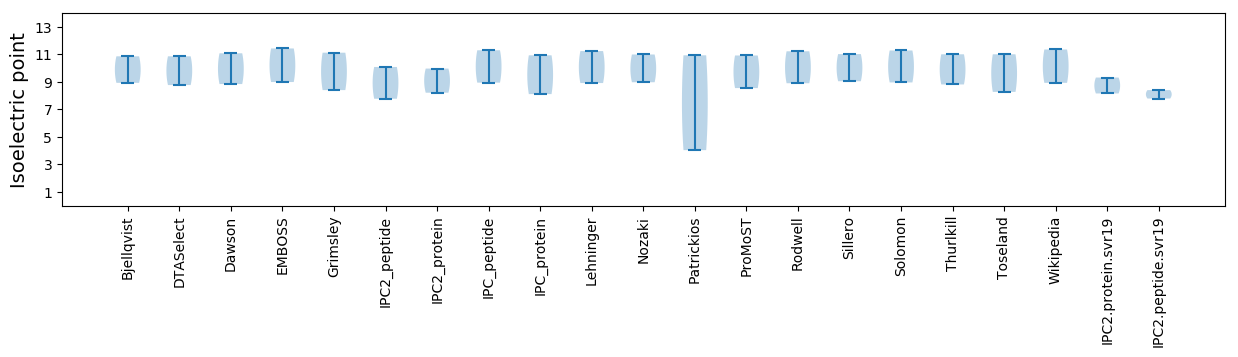
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4N3P7|D4N3P7_9CIRC Capsid protein OS=Cyclovirus Chimp11 OX=742920 GN=cap PE=4 SV=1
MM1 pKa = 7.47AFKK4 pKa = 10.5RR5 pKa = 11.84YY6 pKa = 7.66FRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84VRR14 pKa = 11.84KK15 pKa = 7.51PVRR18 pKa = 11.84RR19 pKa = 11.84FRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.92RR25 pKa = 11.84MRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84SKK34 pKa = 9.88PGNMLTKK41 pKa = 9.51LTKK44 pKa = 9.08ITTLSVEE51 pKa = 4.15NNINATWSCSFKK63 pKa = 10.68MGDD66 pKa = 3.14FTEE69 pKa = 4.29YY70 pKa = 11.04GRR72 pKa = 11.84LAPNFEE78 pKa = 4.37TVKK81 pKa = 10.39LNKK84 pKa = 9.75VVVRR88 pKa = 11.84VQPLQNVANNSTSSVPAYY106 pKa = 10.49VVVPWHH112 pKa = 5.92YY113 pKa = 11.25NIALPKK119 pKa = 10.63DD120 pKa = 3.31FASYY124 pKa = 11.01LRR126 pKa = 11.84IDD128 pKa = 3.44KK129 pKa = 10.75HH130 pKa = 6.08KK131 pKa = 10.92LRR133 pKa = 11.84AQTVGTSMSFVPNIVTVGVANEE155 pKa = 4.2GANPTGRR162 pKa = 11.84NITWKK167 pKa = 8.77PTLEE171 pKa = 4.07CLGVDD176 pKa = 3.18INIPRR181 pKa = 11.84VYY183 pKa = 10.34CGAICFQGQPDD194 pKa = 3.62MEE196 pKa = 4.31GRR198 pKa = 11.84KK199 pKa = 7.26TAFNIITDD207 pKa = 3.78VYY209 pKa = 10.41CTFRR213 pKa = 11.84NQNTMKK219 pKa = 10.53VV220 pKa = 3.37
MM1 pKa = 7.47AFKK4 pKa = 10.5RR5 pKa = 11.84YY6 pKa = 7.66FRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84VRR14 pKa = 11.84KK15 pKa = 7.51PVRR18 pKa = 11.84RR19 pKa = 11.84FRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.92RR25 pKa = 11.84MRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84SKK34 pKa = 9.88PGNMLTKK41 pKa = 9.51LTKK44 pKa = 9.08ITTLSVEE51 pKa = 4.15NNINATWSCSFKK63 pKa = 10.68MGDD66 pKa = 3.14FTEE69 pKa = 4.29YY70 pKa = 11.04GRR72 pKa = 11.84LAPNFEE78 pKa = 4.37TVKK81 pKa = 10.39LNKK84 pKa = 9.75VVVRR88 pKa = 11.84VQPLQNVANNSTSSVPAYY106 pKa = 10.49VVVPWHH112 pKa = 5.92YY113 pKa = 11.25NIALPKK119 pKa = 10.63DD120 pKa = 3.31FASYY124 pKa = 11.01LRR126 pKa = 11.84IDD128 pKa = 3.44KK129 pKa = 10.75HH130 pKa = 6.08KK131 pKa = 10.92LRR133 pKa = 11.84AQTVGTSMSFVPNIVTVGVANEE155 pKa = 4.2GANPTGRR162 pKa = 11.84NITWKK167 pKa = 8.77PTLEE171 pKa = 4.07CLGVDD176 pKa = 3.18INIPRR181 pKa = 11.84VYY183 pKa = 10.34CGAICFQGQPDD194 pKa = 3.62MEE196 pKa = 4.31GRR198 pKa = 11.84KK199 pKa = 7.26TAFNIITDD207 pKa = 3.78VYY209 pKa = 10.41CTFRR213 pKa = 11.84NQNTMKK219 pKa = 10.53VV220 pKa = 3.37
Molecular weight: 25.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
500 |
220 |
280 |
250.0 |
29.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.4 ± 0.693 | 2.2 ± 0.048 |
3.6 ± 0.574 | 5.0 ± 1.495 |
5.0 ± 0.0 | 5.8 ± 0.526 |
1.8 ± 0.586 | 7.0 ± 1.315 |
7.0 ± 0.418 | 5.6 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.777 | 6.8 ± 0.909 |
5.2 ± 0.167 | 3.6 ± 0.574 |
8.4 ± 1.949 | 4.6 ± 0.036 |
8.0 ± 0.12 | 6.6 ± 2.535 |
1.8 ± 0.287 | 5.6 ± 1.291 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
