
Candidatus Kentron sp. G
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Candidatus Kentron; unclassified Candidatus Kentron
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
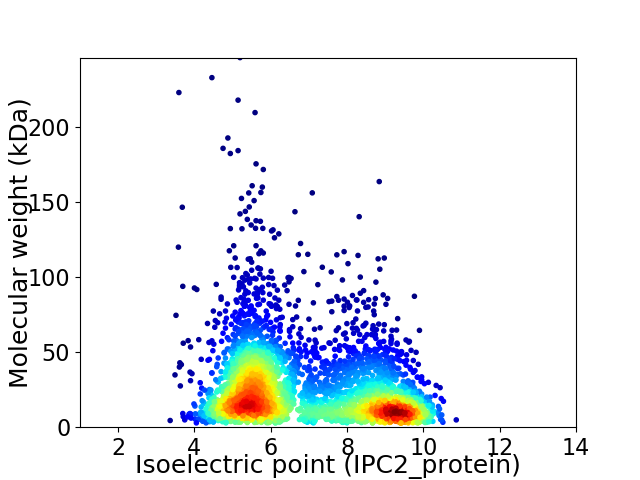
Virtual 2D-PAGE plot for 3885 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A451C6X3|A0A451C6X3_9GAMM Precorrin-2 C20-methyltransferase /cobalt-factor II C20-methyltransferase OS=Candidatus Kentron sp. G OX=2126341 GN=BECKG1743F_GA0114225_101364 PE=3 SV=1
MM1 pKa = 7.58TYY3 pKa = 10.87KK4 pKa = 10.57EE5 pKa = 4.44MNSADD10 pKa = 3.76LGRR13 pKa = 11.84GRR15 pKa = 11.84IAGGYY20 pKa = 9.68FGLLPLEE27 pKa = 4.05SRR29 pKa = 11.84IMFDD33 pKa = 3.15ASAATTVSDD42 pKa = 3.47TGSDD46 pKa = 3.12NGASNTTTASEE57 pKa = 4.29SSSPSSSGEE66 pKa = 3.86VFAEE70 pKa = 4.21SSTKK74 pKa = 10.21EE75 pKa = 3.92SSSEE79 pKa = 3.72EE80 pKa = 3.99GATIISDD87 pKa = 3.63PSSYY91 pKa = 10.8SYY93 pKa = 11.1EE94 pKa = 4.09EE95 pKa = 3.72QAGALAIDD103 pKa = 5.81GITVSTNLDD112 pKa = 3.35YY113 pKa = 11.4HH114 pKa = 6.64GGSVSFTLDD123 pKa = 2.92EE124 pKa = 4.49STSSDD129 pKa = 3.14QLALIFEE136 pKa = 5.1PGDD139 pKa = 3.51PNAEE143 pKa = 4.15GEE145 pKa = 4.32ISLGDD150 pKa = 3.84DD151 pKa = 4.72GISVYY156 pKa = 10.51CGNGSGQEE164 pKa = 4.0LIGWIDD170 pKa = 3.58SDD172 pKa = 4.77NNGQNGTSLQINFRR186 pKa = 11.84EE187 pKa = 4.1DD188 pKa = 2.83QFANSTFEE196 pKa = 4.17NDD198 pKa = 4.37GEE200 pKa = 4.28ATLNQGLEE208 pKa = 3.9EE209 pKa = 5.31DD210 pKa = 3.58GWILQKK216 pKa = 10.91EE217 pKa = 4.23SWTYY221 pKa = 11.14SSALGAYY228 pKa = 8.62GPDD231 pKa = 2.85MSVIFEE237 pKa = 4.53GPSADD242 pKa = 4.5RR243 pKa = 11.84ICVSDD248 pKa = 3.9TEE250 pKa = 5.26CYY252 pKa = 10.24DD253 pKa = 3.4IPYY256 pKa = 9.89LDD258 pKa = 4.3YY259 pKa = 11.32EE260 pKa = 4.5EE261 pKa = 5.72PPDD264 pKa = 3.55STEE267 pKa = 3.74YY268 pKa = 10.61LGQGFSPPVGEE279 pKa = 4.24WLIGDD284 pKa = 3.76QYY286 pKa = 11.41ISGDD290 pKa = 3.04TWINIDD296 pKa = 4.48DD297 pKa = 4.18NEE299 pKa = 4.17SSYY302 pKa = 11.77VYY304 pKa = 10.97ALNEE308 pKa = 4.01SDD310 pKa = 3.72TFLLGSQGVVMDD322 pKa = 3.92SAFNGEE328 pKa = 4.54SVVMHH333 pKa = 6.5GPYY336 pKa = 10.28ACSNEE341 pKa = 3.89VGLIEE346 pKa = 5.06GDD348 pKa = 3.64TVRR351 pKa = 11.84FDD353 pKa = 3.28WKK355 pKa = 10.67GEE357 pKa = 3.69EE358 pKa = 4.0RR359 pKa = 11.84FEE361 pKa = 4.44GDD363 pKa = 3.34EE364 pKa = 4.24YY365 pKa = 11.83VVFAYY370 pKa = 10.55LLKK373 pKa = 10.11TDD375 pKa = 4.5GEE377 pKa = 4.61NAGKK381 pKa = 8.11TKK383 pKa = 10.8VMLNEE388 pKa = 4.06VGGGDD393 pKa = 3.67DD394 pKa = 4.81NEE396 pKa = 4.41WKK398 pKa = 9.14TVEE401 pKa = 4.19TTISAGEE408 pKa = 4.29DD409 pKa = 2.98GTYY412 pKa = 10.11RR413 pKa = 11.84AVFISGSIDD422 pKa = 3.15TSGGGLVGAVFHH434 pKa = 7.15LDD436 pKa = 3.19NLMLGPSIDD445 pKa = 3.66GTVLTNIASGLEE457 pKa = 4.0FSNDD461 pKa = 3.06SYY463 pKa = 12.07DD464 pKa = 3.51PGEE467 pKa = 3.99IRR469 pKa = 11.84EE470 pKa = 4.29LTISVQSSDD479 pKa = 3.6KK480 pKa = 11.0TDD482 pKa = 4.36DD483 pKa = 4.42PDD485 pKa = 3.84TQTVSIDD492 pKa = 3.1IEE494 pKa = 4.39QKK496 pKa = 10.52NDD498 pKa = 3.43APVLTADD505 pKa = 4.86GEE507 pKa = 4.48ITLEE511 pKa = 4.08AGEE514 pKa = 4.17VSEE517 pKa = 4.25LSGVRR522 pKa = 11.84VEE524 pKa = 4.73DD525 pKa = 3.88ADD527 pKa = 3.78VGEE530 pKa = 4.66GEE532 pKa = 4.09IQIEE536 pKa = 4.27ITVDD540 pKa = 3.49DD541 pKa = 4.34PAVGVLQVSEE551 pKa = 5.4DD552 pKa = 3.74YY553 pKa = 11.59AEE555 pKa = 5.38LITGGGNNSATVTLEE570 pKa = 3.82GTLDD574 pKa = 3.72EE575 pKa = 4.92VNSALNTLAFVAEE588 pKa = 4.63SEE590 pKa = 4.54AEE592 pKa = 4.04PGTTQTQIQYY602 pKa = 10.55SVSDD606 pKa = 4.1LGNTGATAEE615 pKa = 4.22TAYY618 pKa = 10.78GEE620 pKa = 4.37TTVFVDD626 pKa = 4.53GGDD629 pKa = 3.81PVDD632 pKa = 4.91GGDD635 pKa = 4.14PVDD638 pKa = 4.91GGDD641 pKa = 4.14PVDD644 pKa = 4.66GGDD647 pKa = 3.73PGLVGSGPDD656 pKa = 3.23RR657 pKa = 11.84TGSLYY662 pKa = 10.69SDD664 pKa = 4.96PYY666 pKa = 11.09SMSSVTSSLGTSVAAVYY683 pKa = 10.34RR684 pKa = 11.84DD685 pKa = 3.81SEE687 pKa = 5.09CNFPATPSCLFYY699 pKa = 10.97FRR701 pKa = 11.84DD702 pKa = 3.39YY703 pKa = 11.82GLIHH707 pKa = 5.85EE708 pKa = 5.03TEE710 pKa = 4.35NGTSGAGEE718 pKa = 3.8AFAFQLPYY726 pKa = 10.59DD727 pKa = 4.09ALIHH731 pKa = 5.67TSYY734 pKa = 10.61HH735 pKa = 5.39YY736 pKa = 10.75YY737 pKa = 10.39CRR739 pKa = 11.84EE740 pKa = 4.16TQWDD744 pKa = 3.89TRR746 pKa = 11.84QLDD749 pKa = 4.05GSPLPRR755 pKa = 11.84WLDD758 pKa = 3.57FNPEE762 pKa = 3.95KK763 pKa = 10.72GTVTGTAPEE772 pKa = 4.24YY773 pKa = 10.56LAGTVVEE780 pKa = 4.78LEE782 pKa = 4.56FIARR786 pKa = 11.84DD787 pKa = 3.45VSSQQEE793 pKa = 3.95IAVLRR798 pKa = 11.84FNLCVVEE805 pKa = 6.18DD806 pKa = 4.3RR807 pKa = 11.84AQEE810 pKa = 4.11CQPKK814 pKa = 10.18LVEE817 pKa = 4.24EE818 pKa = 4.24KK819 pKa = 10.56QGNDD823 pKa = 3.52EE824 pKa = 4.7LSHH827 pKa = 7.23ADD829 pKa = 3.08TDD831 pKa = 3.44IDD833 pKa = 4.22FRR835 pKa = 11.84NNMPPSAHH843 pKa = 5.37TVQNISRR850 pKa = 11.84PAFSAQLAAAGRR862 pKa = 11.84AGFEE866 pKa = 3.67ARR868 pKa = 11.84QAAFITMLGSHH879 pKa = 6.7GLL881 pKa = 3.53
MM1 pKa = 7.58TYY3 pKa = 10.87KK4 pKa = 10.57EE5 pKa = 4.44MNSADD10 pKa = 3.76LGRR13 pKa = 11.84GRR15 pKa = 11.84IAGGYY20 pKa = 9.68FGLLPLEE27 pKa = 4.05SRR29 pKa = 11.84IMFDD33 pKa = 3.15ASAATTVSDD42 pKa = 3.47TGSDD46 pKa = 3.12NGASNTTTASEE57 pKa = 4.29SSSPSSSGEE66 pKa = 3.86VFAEE70 pKa = 4.21SSTKK74 pKa = 10.21EE75 pKa = 3.92SSSEE79 pKa = 3.72EE80 pKa = 3.99GATIISDD87 pKa = 3.63PSSYY91 pKa = 10.8SYY93 pKa = 11.1EE94 pKa = 4.09EE95 pKa = 3.72QAGALAIDD103 pKa = 5.81GITVSTNLDD112 pKa = 3.35YY113 pKa = 11.4HH114 pKa = 6.64GGSVSFTLDD123 pKa = 2.92EE124 pKa = 4.49STSSDD129 pKa = 3.14QLALIFEE136 pKa = 5.1PGDD139 pKa = 3.51PNAEE143 pKa = 4.15GEE145 pKa = 4.32ISLGDD150 pKa = 3.84DD151 pKa = 4.72GISVYY156 pKa = 10.51CGNGSGQEE164 pKa = 4.0LIGWIDD170 pKa = 3.58SDD172 pKa = 4.77NNGQNGTSLQINFRR186 pKa = 11.84EE187 pKa = 4.1DD188 pKa = 2.83QFANSTFEE196 pKa = 4.17NDD198 pKa = 4.37GEE200 pKa = 4.28ATLNQGLEE208 pKa = 3.9EE209 pKa = 5.31DD210 pKa = 3.58GWILQKK216 pKa = 10.91EE217 pKa = 4.23SWTYY221 pKa = 11.14SSALGAYY228 pKa = 8.62GPDD231 pKa = 2.85MSVIFEE237 pKa = 4.53GPSADD242 pKa = 4.5RR243 pKa = 11.84ICVSDD248 pKa = 3.9TEE250 pKa = 5.26CYY252 pKa = 10.24DD253 pKa = 3.4IPYY256 pKa = 9.89LDD258 pKa = 4.3YY259 pKa = 11.32EE260 pKa = 4.5EE261 pKa = 5.72PPDD264 pKa = 3.55STEE267 pKa = 3.74YY268 pKa = 10.61LGQGFSPPVGEE279 pKa = 4.24WLIGDD284 pKa = 3.76QYY286 pKa = 11.41ISGDD290 pKa = 3.04TWINIDD296 pKa = 4.48DD297 pKa = 4.18NEE299 pKa = 4.17SSYY302 pKa = 11.77VYY304 pKa = 10.97ALNEE308 pKa = 4.01SDD310 pKa = 3.72TFLLGSQGVVMDD322 pKa = 3.92SAFNGEE328 pKa = 4.54SVVMHH333 pKa = 6.5GPYY336 pKa = 10.28ACSNEE341 pKa = 3.89VGLIEE346 pKa = 5.06GDD348 pKa = 3.64TVRR351 pKa = 11.84FDD353 pKa = 3.28WKK355 pKa = 10.67GEE357 pKa = 3.69EE358 pKa = 4.0RR359 pKa = 11.84FEE361 pKa = 4.44GDD363 pKa = 3.34EE364 pKa = 4.24YY365 pKa = 11.83VVFAYY370 pKa = 10.55LLKK373 pKa = 10.11TDD375 pKa = 4.5GEE377 pKa = 4.61NAGKK381 pKa = 8.11TKK383 pKa = 10.8VMLNEE388 pKa = 4.06VGGGDD393 pKa = 3.67DD394 pKa = 4.81NEE396 pKa = 4.41WKK398 pKa = 9.14TVEE401 pKa = 4.19TTISAGEE408 pKa = 4.29DD409 pKa = 2.98GTYY412 pKa = 10.11RR413 pKa = 11.84AVFISGSIDD422 pKa = 3.15TSGGGLVGAVFHH434 pKa = 7.15LDD436 pKa = 3.19NLMLGPSIDD445 pKa = 3.66GTVLTNIASGLEE457 pKa = 4.0FSNDD461 pKa = 3.06SYY463 pKa = 12.07DD464 pKa = 3.51PGEE467 pKa = 3.99IRR469 pKa = 11.84EE470 pKa = 4.29LTISVQSSDD479 pKa = 3.6KK480 pKa = 11.0TDD482 pKa = 4.36DD483 pKa = 4.42PDD485 pKa = 3.84TQTVSIDD492 pKa = 3.1IEE494 pKa = 4.39QKK496 pKa = 10.52NDD498 pKa = 3.43APVLTADD505 pKa = 4.86GEE507 pKa = 4.48ITLEE511 pKa = 4.08AGEE514 pKa = 4.17VSEE517 pKa = 4.25LSGVRR522 pKa = 11.84VEE524 pKa = 4.73DD525 pKa = 3.88ADD527 pKa = 3.78VGEE530 pKa = 4.66GEE532 pKa = 4.09IQIEE536 pKa = 4.27ITVDD540 pKa = 3.49DD541 pKa = 4.34PAVGVLQVSEE551 pKa = 5.4DD552 pKa = 3.74YY553 pKa = 11.59AEE555 pKa = 5.38LITGGGNNSATVTLEE570 pKa = 3.82GTLDD574 pKa = 3.72EE575 pKa = 4.92VNSALNTLAFVAEE588 pKa = 4.63SEE590 pKa = 4.54AEE592 pKa = 4.04PGTTQTQIQYY602 pKa = 10.55SVSDD606 pKa = 4.1LGNTGATAEE615 pKa = 4.22TAYY618 pKa = 10.78GEE620 pKa = 4.37TTVFVDD626 pKa = 4.53GGDD629 pKa = 3.81PVDD632 pKa = 4.91GGDD635 pKa = 4.14PVDD638 pKa = 4.91GGDD641 pKa = 4.14PVDD644 pKa = 4.66GGDD647 pKa = 3.73PGLVGSGPDD656 pKa = 3.23RR657 pKa = 11.84TGSLYY662 pKa = 10.69SDD664 pKa = 4.96PYY666 pKa = 11.09SMSSVTSSLGTSVAAVYY683 pKa = 10.34RR684 pKa = 11.84DD685 pKa = 3.81SEE687 pKa = 5.09CNFPATPSCLFYY699 pKa = 10.97FRR701 pKa = 11.84DD702 pKa = 3.39YY703 pKa = 11.82GLIHH707 pKa = 5.85EE708 pKa = 5.03TEE710 pKa = 4.35NGTSGAGEE718 pKa = 3.8AFAFQLPYY726 pKa = 10.59DD727 pKa = 4.09ALIHH731 pKa = 5.67TSYY734 pKa = 10.61HH735 pKa = 5.39YY736 pKa = 10.75YY737 pKa = 10.39CRR739 pKa = 11.84EE740 pKa = 4.16TQWDD744 pKa = 3.89TRR746 pKa = 11.84QLDD749 pKa = 4.05GSPLPRR755 pKa = 11.84WLDD758 pKa = 3.57FNPEE762 pKa = 3.95KK763 pKa = 10.72GTVTGTAPEE772 pKa = 4.24YY773 pKa = 10.56LAGTVVEE780 pKa = 4.78LEE782 pKa = 4.56FIARR786 pKa = 11.84DD787 pKa = 3.45VSSQQEE793 pKa = 3.95IAVLRR798 pKa = 11.84FNLCVVEE805 pKa = 6.18DD806 pKa = 4.3RR807 pKa = 11.84AQEE810 pKa = 4.11CQPKK814 pKa = 10.18LVEE817 pKa = 4.24EE818 pKa = 4.24KK819 pKa = 10.56QGNDD823 pKa = 3.52EE824 pKa = 4.7LSHH827 pKa = 7.23ADD829 pKa = 3.08TDD831 pKa = 3.44IDD833 pKa = 4.22FRR835 pKa = 11.84NNMPPSAHH843 pKa = 5.37TVQNISRR850 pKa = 11.84PAFSAQLAAAGRR862 pKa = 11.84AGFEE866 pKa = 3.67ARR868 pKa = 11.84QAAFITMLGSHH879 pKa = 6.7GLL881 pKa = 3.53
Molecular weight: 93.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A451CCD8|A0A451CCD8_9GAMM Uncharacterized protein OS=Candidatus Kentron sp. G OX=2126341 GN=BECKG1743F_GA0114225_105832 PE=4 SV=1
MM1 pKa = 7.43VHH3 pKa = 6.15EE4 pKa = 4.98HH5 pKa = 7.21PIAHH9 pKa = 6.28TAPRR13 pKa = 11.84QRR15 pKa = 11.84LHH17 pKa = 6.7RR18 pKa = 11.84WFSCPSGKK26 pKa = 10.31ALLDD30 pKa = 3.67RR31 pKa = 11.84EE32 pKa = 4.41RR33 pKa = 11.84KK34 pKa = 9.94LLDD37 pKa = 3.43GVLPHH42 pKa = 7.1LFGHH46 pKa = 6.94YY47 pKa = 9.25ILQVGRR53 pKa = 11.84LGDD56 pKa = 3.92ADD58 pKa = 4.01LLARR62 pKa = 11.84SRR64 pKa = 11.84IPHH67 pKa = 5.34RR68 pKa = 11.84VVVEE72 pKa = 3.55VDD74 pKa = 3.16GNRR77 pKa = 11.84NVSGYY82 pKa = 9.42PRR84 pKa = 11.84LRR86 pKa = 11.84AEE88 pKa = 3.9PHH90 pKa = 6.04ALPVASDD97 pKa = 3.61SVDD100 pKa = 3.79VVILPHH106 pKa = 7.34LLEE109 pKa = 5.44FSAHH113 pKa = 4.5MQATIQEE120 pKa = 4.31AKK122 pKa = 10.09RR123 pKa = 11.84ILVAEE128 pKa = 4.11GHH130 pKa = 6.35LLILAFNAWSLVGIQCLLCRR150 pKa = 11.84ARR152 pKa = 11.84HH153 pKa = 5.02VDD155 pKa = 4.1RR156 pKa = 11.84GTDD159 pKa = 3.1SCLPCPGTRR168 pKa = 11.84FPGINRR174 pKa = 11.84IKK176 pKa = 10.8NHH178 pKa = 5.66IAVLGFDD185 pKa = 4.37IISIDD190 pKa = 3.33RR191 pKa = 11.84YY192 pKa = 9.66FFQPAVANPALMDD205 pKa = 3.86RR206 pKa = 11.84LRR208 pKa = 11.84FLDD211 pKa = 3.46VMGPRR216 pKa = 11.84FWPLLSGAYY225 pKa = 9.9FIVAKK230 pKa = 10.56KK231 pKa = 10.18RR232 pKa = 11.84VTTLTPIKK240 pKa = 10.21LRR242 pKa = 11.84RR243 pKa = 11.84QQPRR247 pKa = 11.84RR248 pKa = 11.84FLIAAGLGEE257 pKa = 4.49SSSCRR262 pKa = 11.84QASEE266 pKa = 3.71RR267 pKa = 11.84RR268 pKa = 11.84PRR270 pKa = 3.69
MM1 pKa = 7.43VHH3 pKa = 6.15EE4 pKa = 4.98HH5 pKa = 7.21PIAHH9 pKa = 6.28TAPRR13 pKa = 11.84QRR15 pKa = 11.84LHH17 pKa = 6.7RR18 pKa = 11.84WFSCPSGKK26 pKa = 10.31ALLDD30 pKa = 3.67RR31 pKa = 11.84EE32 pKa = 4.41RR33 pKa = 11.84KK34 pKa = 9.94LLDD37 pKa = 3.43GVLPHH42 pKa = 7.1LFGHH46 pKa = 6.94YY47 pKa = 9.25ILQVGRR53 pKa = 11.84LGDD56 pKa = 3.92ADD58 pKa = 4.01LLARR62 pKa = 11.84SRR64 pKa = 11.84IPHH67 pKa = 5.34RR68 pKa = 11.84VVVEE72 pKa = 3.55VDD74 pKa = 3.16GNRR77 pKa = 11.84NVSGYY82 pKa = 9.42PRR84 pKa = 11.84LRR86 pKa = 11.84AEE88 pKa = 3.9PHH90 pKa = 6.04ALPVASDD97 pKa = 3.61SVDD100 pKa = 3.79VVILPHH106 pKa = 7.34LLEE109 pKa = 5.44FSAHH113 pKa = 4.5MQATIQEE120 pKa = 4.31AKK122 pKa = 10.09RR123 pKa = 11.84ILVAEE128 pKa = 4.11GHH130 pKa = 6.35LLILAFNAWSLVGIQCLLCRR150 pKa = 11.84ARR152 pKa = 11.84HH153 pKa = 5.02VDD155 pKa = 4.1RR156 pKa = 11.84GTDD159 pKa = 3.1SCLPCPGTRR168 pKa = 11.84FPGINRR174 pKa = 11.84IKK176 pKa = 10.8NHH178 pKa = 5.66IAVLGFDD185 pKa = 4.37IISIDD190 pKa = 3.33RR191 pKa = 11.84YY192 pKa = 9.66FFQPAVANPALMDD205 pKa = 3.86RR206 pKa = 11.84LRR208 pKa = 11.84FLDD211 pKa = 3.46VMGPRR216 pKa = 11.84FWPLLSGAYY225 pKa = 9.9FIVAKK230 pKa = 10.56KK231 pKa = 10.18RR232 pKa = 11.84VTTLTPIKK240 pKa = 10.21LRR242 pKa = 11.84RR243 pKa = 11.84QQPRR247 pKa = 11.84RR248 pKa = 11.84FLIAAGLGEE257 pKa = 4.49SSSCRR262 pKa = 11.84QASEE266 pKa = 3.71RR267 pKa = 11.84RR268 pKa = 11.84PRR270 pKa = 3.69
Molecular weight: 30.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952963 |
24 |
2253 |
245.3 |
27.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.442 ± 0.048 | 1.155 ± 0.018 |
5.56 ± 0.041 | 6.488 ± 0.041 |
3.971 ± 0.03 | 8.07 ± 0.042 |
2.46 ± 0.021 | 5.791 ± 0.035 |
3.953 ± 0.036 | 10.121 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.021 | 3.144 ± 0.023 |
5.188 ± 0.035 | 3.254 ± 0.028 |
7.6 ± 0.045 | 5.551 ± 0.029 |
5.17 ± 0.039 | 6.564 ± 0.033 |
1.371 ± 0.02 | 2.783 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
