
Capybara microvirus Cap3_SP_470
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 4.82
Get precalculated fractions of proteins
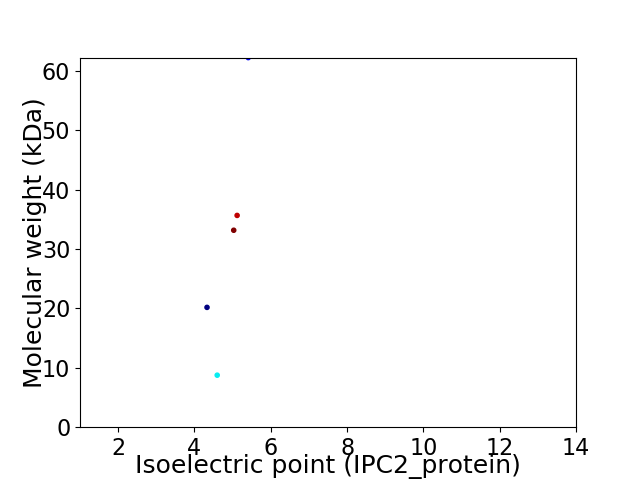
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
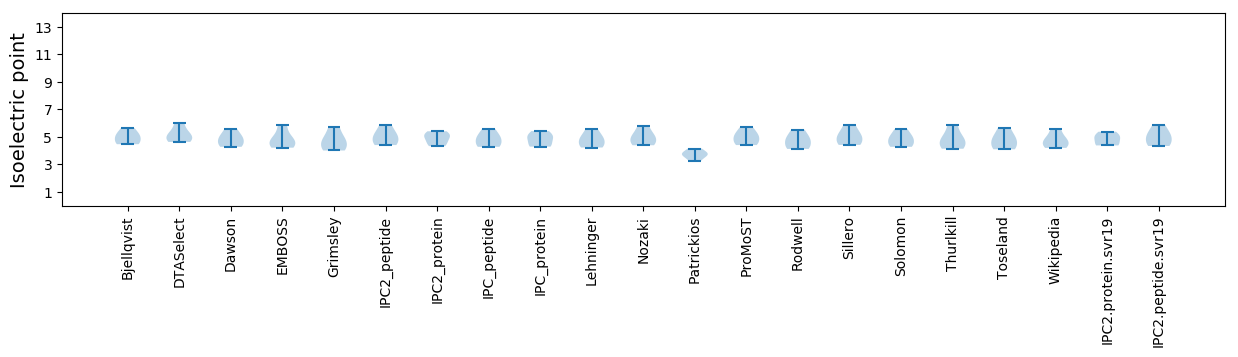
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5A5|A0A4P8W5A5_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_470 OX=2585465 PE=4 SV=1
MM1 pKa = 7.79IYY3 pKa = 10.49GIYY6 pKa = 10.07CMKK9 pKa = 10.48DD10 pKa = 3.09VKK12 pKa = 10.24TGFLNPFVEE21 pKa = 4.49VNDD24 pKa = 3.97DD25 pKa = 3.47VAVRR29 pKa = 11.84GFANAMMAPGTVYY42 pKa = 11.17SNFSADD48 pKa = 3.26FSLYY52 pKa = 10.43KK53 pKa = 10.62VGLFDD58 pKa = 5.96SEE60 pKa = 4.99LCVLSGVDD68 pKa = 3.12KK69 pKa = 11.35VLIAEE74 pKa = 4.9ASQFVKK80 pKa = 10.89
MM1 pKa = 7.79IYY3 pKa = 10.49GIYY6 pKa = 10.07CMKK9 pKa = 10.48DD10 pKa = 3.09VKK12 pKa = 10.24TGFLNPFVEE21 pKa = 4.49VNDD24 pKa = 3.97DD25 pKa = 3.47VAVRR29 pKa = 11.84GFANAMMAPGTVYY42 pKa = 11.17SNFSADD48 pKa = 3.26FSLYY52 pKa = 10.43KK53 pKa = 10.62VGLFDD58 pKa = 5.96SEE60 pKa = 4.99LCVLSGVDD68 pKa = 3.12KK69 pKa = 11.35VLIAEE74 pKa = 4.9ASQFVKK80 pKa = 10.89
Molecular weight: 8.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVU1|A0A4V1FVU1_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_470 OX=2585465 PE=4 SV=1
MM1 pKa = 7.49NSRR4 pKa = 11.84FAVAPQVNINRR15 pKa = 11.84STFRR19 pKa = 11.84RR20 pKa = 11.84DD21 pKa = 3.17HH22 pKa = 5.11TVKK25 pKa = 10.05TSFNMGEE32 pKa = 4.66LIPFCVEE39 pKa = 3.48EE40 pKa = 4.47VLPGDD45 pKa = 3.63TFSIKK50 pKa = 8.91TSKK53 pKa = 10.8LIRR56 pKa = 11.84TQPLLTPLMDD66 pKa = 3.69QLYY69 pKa = 10.75LDD71 pKa = 4.24TYY73 pKa = 10.35WYY75 pKa = 9.69FVPNRR80 pKa = 11.84LIWEE84 pKa = 3.82HH85 pKa = 6.93WINFMGEE92 pKa = 4.08NTDD95 pKa = 3.7GSWIPEE101 pKa = 4.08TQYY104 pKa = 11.57SVPTVSAPSSTGWIEE119 pKa = 3.9HH120 pKa = 5.93TLADD124 pKa = 4.48YY125 pKa = 9.9MGLPVNVPSSSDD137 pKa = 3.31GVFSVSALPFRR148 pKa = 11.84AYY150 pKa = 10.59CKK152 pKa = 10.05IYY154 pKa = 10.65NDD156 pKa = 3.35WFRR159 pKa = 11.84DD160 pKa = 3.61EE161 pKa = 4.93NLCDD165 pKa = 3.72PAFLPLDD172 pKa = 3.89DD173 pKa = 4.02STRR176 pKa = 11.84VGSNGDD182 pKa = 3.7SLNDD186 pKa = 3.31VCLGGMPLKK195 pKa = 10.48SGKK198 pKa = 9.12IHH200 pKa = 7.77DD201 pKa = 4.59YY202 pKa = 10.78FSSCLPAPQKK212 pKa = 10.77GSPVSLGLEE221 pKa = 4.04SVYY224 pKa = 11.16GDD226 pKa = 3.06GFVPVFGNGLSLGLHH241 pKa = 5.12NSEE244 pKa = 4.39RR245 pKa = 11.84LFGISSSNSSGLLGASSNAVGNPLGTTTSSIVPIQNKK282 pKa = 10.25SIGVLTKK289 pKa = 10.59SGLDD293 pKa = 3.54AVGLPYY299 pKa = 10.51KK300 pKa = 10.1DD301 pKa = 3.41TGLVADD307 pKa = 4.81VSQSSLTVNSLRR319 pKa = 11.84YY320 pKa = 9.36AFAVQRR326 pKa = 11.84LLEE329 pKa = 4.37KK330 pKa = 10.4DD331 pKa = 3.05ARR333 pKa = 11.84GGSRR337 pKa = 11.84YY338 pKa = 9.2IEE340 pKa = 3.95QIKK343 pKa = 9.88SHH345 pKa = 6.73FGVTSPDD352 pKa = 2.98ARR354 pKa = 11.84LQRR357 pKa = 11.84SEE359 pKa = 4.06YY360 pKa = 10.84LGGNRR365 pKa = 11.84INININQVLNVAQAQKK381 pKa = 9.02TGSSEE386 pKa = 4.0GMLGNVAGYY395 pKa = 10.35SVTGDD400 pKa = 3.3SHH402 pKa = 8.24GDD404 pKa = 3.34FSKK407 pKa = 11.1SFTEE411 pKa = 3.58HH412 pKa = 6.5GYY414 pKa = 10.25IIGLMTVRR422 pKa = 11.84QDD424 pKa = 3.12NSYY427 pKa = 11.04CQGTDD432 pKa = 3.19RR433 pKa = 11.84FWKK436 pKa = 9.77RR437 pKa = 11.84KK438 pKa = 6.8TRR440 pKa = 11.84FDD442 pKa = 3.24YY443 pKa = 10.8YY444 pKa = 11.31YY445 pKa = 10.13PVFANIGEE453 pKa = 4.24QPVYY457 pKa = 10.31QSEE460 pKa = 4.41IYY462 pKa = 8.31EE463 pKa = 4.3TGSNSDD469 pKa = 2.83IVFGYY474 pKa = 9.6QEE476 pKa = 3.41AWADD480 pKa = 3.69YY481 pKa = 9.96RR482 pKa = 11.84FKK484 pKa = 10.56PSKK487 pKa = 8.03VTGMMRR493 pKa = 11.84NQLDD497 pKa = 3.26AWHH500 pKa = 6.86LADD503 pKa = 5.24KK504 pKa = 10.84YY505 pKa = 11.33NNPPVLGKK513 pKa = 8.1TWIEE517 pKa = 3.7SDD519 pKa = 3.46KK520 pKa = 11.65ANLDD524 pKa = 3.25RR525 pKa = 11.84ALAVTSSTADD535 pKa = 3.04QFFADD540 pKa = 4.04IFIEE544 pKa = 4.34NKK546 pKa = 8.05CTRR549 pKa = 11.84PMPMFSIPGLLDD561 pKa = 3.34HH562 pKa = 6.99NN563 pKa = 4.67
MM1 pKa = 7.49NSRR4 pKa = 11.84FAVAPQVNINRR15 pKa = 11.84STFRR19 pKa = 11.84RR20 pKa = 11.84DD21 pKa = 3.17HH22 pKa = 5.11TVKK25 pKa = 10.05TSFNMGEE32 pKa = 4.66LIPFCVEE39 pKa = 3.48EE40 pKa = 4.47VLPGDD45 pKa = 3.63TFSIKK50 pKa = 8.91TSKK53 pKa = 10.8LIRR56 pKa = 11.84TQPLLTPLMDD66 pKa = 3.69QLYY69 pKa = 10.75LDD71 pKa = 4.24TYY73 pKa = 10.35WYY75 pKa = 9.69FVPNRR80 pKa = 11.84LIWEE84 pKa = 3.82HH85 pKa = 6.93WINFMGEE92 pKa = 4.08NTDD95 pKa = 3.7GSWIPEE101 pKa = 4.08TQYY104 pKa = 11.57SVPTVSAPSSTGWIEE119 pKa = 3.9HH120 pKa = 5.93TLADD124 pKa = 4.48YY125 pKa = 9.9MGLPVNVPSSSDD137 pKa = 3.31GVFSVSALPFRR148 pKa = 11.84AYY150 pKa = 10.59CKK152 pKa = 10.05IYY154 pKa = 10.65NDD156 pKa = 3.35WFRR159 pKa = 11.84DD160 pKa = 3.61EE161 pKa = 4.93NLCDD165 pKa = 3.72PAFLPLDD172 pKa = 3.89DD173 pKa = 4.02STRR176 pKa = 11.84VGSNGDD182 pKa = 3.7SLNDD186 pKa = 3.31VCLGGMPLKK195 pKa = 10.48SGKK198 pKa = 9.12IHH200 pKa = 7.77DD201 pKa = 4.59YY202 pKa = 10.78FSSCLPAPQKK212 pKa = 10.77GSPVSLGLEE221 pKa = 4.04SVYY224 pKa = 11.16GDD226 pKa = 3.06GFVPVFGNGLSLGLHH241 pKa = 5.12NSEE244 pKa = 4.39RR245 pKa = 11.84LFGISSSNSSGLLGASSNAVGNPLGTTTSSIVPIQNKK282 pKa = 10.25SIGVLTKK289 pKa = 10.59SGLDD293 pKa = 3.54AVGLPYY299 pKa = 10.51KK300 pKa = 10.1DD301 pKa = 3.41TGLVADD307 pKa = 4.81VSQSSLTVNSLRR319 pKa = 11.84YY320 pKa = 9.36AFAVQRR326 pKa = 11.84LLEE329 pKa = 4.37KK330 pKa = 10.4DD331 pKa = 3.05ARR333 pKa = 11.84GGSRR337 pKa = 11.84YY338 pKa = 9.2IEE340 pKa = 3.95QIKK343 pKa = 9.88SHH345 pKa = 6.73FGVTSPDD352 pKa = 2.98ARR354 pKa = 11.84LQRR357 pKa = 11.84SEE359 pKa = 4.06YY360 pKa = 10.84LGGNRR365 pKa = 11.84INININQVLNVAQAQKK381 pKa = 9.02TGSSEE386 pKa = 4.0GMLGNVAGYY395 pKa = 10.35SVTGDD400 pKa = 3.3SHH402 pKa = 8.24GDD404 pKa = 3.34FSKK407 pKa = 11.1SFTEE411 pKa = 3.58HH412 pKa = 6.5GYY414 pKa = 10.25IIGLMTVRR422 pKa = 11.84QDD424 pKa = 3.12NSYY427 pKa = 11.04CQGTDD432 pKa = 3.19RR433 pKa = 11.84FWKK436 pKa = 9.77RR437 pKa = 11.84KK438 pKa = 6.8TRR440 pKa = 11.84FDD442 pKa = 3.24YY443 pKa = 10.8YY444 pKa = 11.31YY445 pKa = 10.13PVFANIGEE453 pKa = 4.24QPVYY457 pKa = 10.31QSEE460 pKa = 4.41IYY462 pKa = 8.31EE463 pKa = 4.3TGSNSDD469 pKa = 2.83IVFGYY474 pKa = 9.6QEE476 pKa = 3.41AWADD480 pKa = 3.69YY481 pKa = 9.96RR482 pKa = 11.84FKK484 pKa = 10.56PSKK487 pKa = 8.03VTGMMRR493 pKa = 11.84NQLDD497 pKa = 3.26AWHH500 pKa = 6.86LADD503 pKa = 5.24KK504 pKa = 10.84YY505 pKa = 11.33NNPPVLGKK513 pKa = 8.1TWIEE517 pKa = 3.7SDD519 pKa = 3.46KK520 pKa = 11.65ANLDD524 pKa = 3.25RR525 pKa = 11.84ALAVTSSTADD535 pKa = 3.04QFFADD540 pKa = 4.04IFIEE544 pKa = 4.34NKK546 pKa = 8.05CTRR549 pKa = 11.84PMPMFSIPGLLDD561 pKa = 3.34HH562 pKa = 6.99NN563 pKa = 4.67
Molecular weight: 62.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1445 |
80 |
563 |
289.0 |
32.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.336 ± 1.792 | 1.038 ± 0.285 |
6.436 ± 1.046 | 5.19 ± 0.756 |
4.498 ± 0.684 | 6.92 ± 0.755 |
1.384 ± 0.414 | 5.19 ± 0.935 |
4.983 ± 0.599 | 8.235 ± 0.97 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.353 ± 0.273 | 6.505 ± 0.439 |
4.014 ± 0.717 | 3.875 ± 0.566 |
3.668 ± 0.534 | 11.003 ± 1.821 |
5.26 ± 0.606 | 6.09 ± 0.754 |
1.522 ± 0.167 | 4.498 ± 0.529 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
