
Paramaledivibacter caminithermalis DSM 15212
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Paramaledivibacter; Paramaledivibacter caminithermalis
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
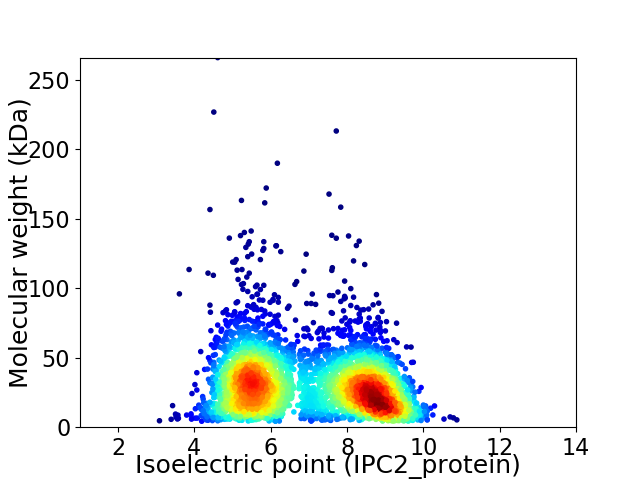
Virtual 2D-PAGE plot for 3784 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
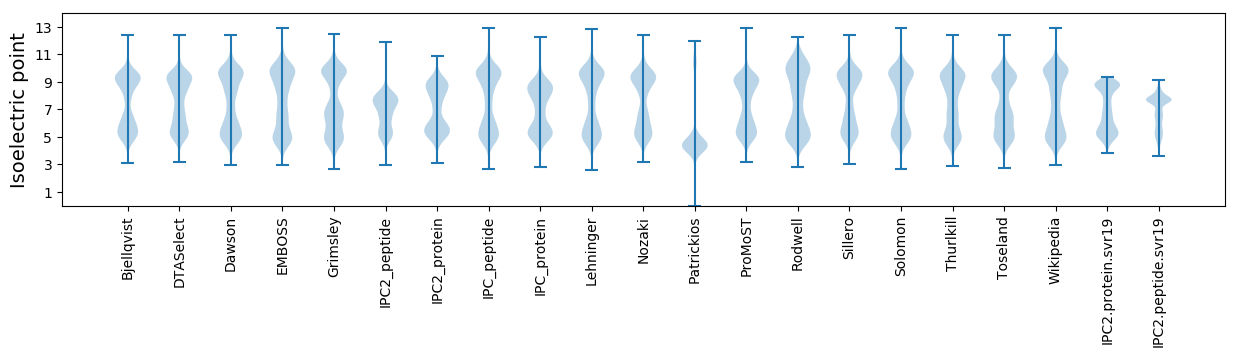
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6Q1N6|A0A1M6Q1N6_9CLOT L-asparaginase OS=Paramaledivibacter caminithermalis DSM 15212 OX=1121301 GN=SAMN02745912_02381 PE=3 SV=1
MM1 pKa = 7.48KK2 pKa = 10.05KK3 pKa = 10.16YY4 pKa = 10.52RR5 pKa = 11.84CIPCGYY11 pKa = 9.88EE12 pKa = 3.57YY13 pKa = 10.65DD14 pKa = 4.1PQVGDD19 pKa = 4.09PDD21 pKa = 4.19SGVAPGTAFEE31 pKa = 5.97DD32 pKa = 4.28IPDD35 pKa = 3.7DD36 pKa = 3.88WVCPVCGVGKK46 pKa = 10.52DD47 pKa = 3.17MFEE50 pKa = 4.09PVEE53 pKa = 4.0
MM1 pKa = 7.48KK2 pKa = 10.05KK3 pKa = 10.16YY4 pKa = 10.52RR5 pKa = 11.84CIPCGYY11 pKa = 9.88EE12 pKa = 3.57YY13 pKa = 10.65DD14 pKa = 4.1PQVGDD19 pKa = 4.09PDD21 pKa = 4.19SGVAPGTAFEE31 pKa = 5.97DD32 pKa = 4.28IPDD35 pKa = 3.7DD36 pKa = 3.88WVCPVCGVGKK46 pKa = 10.52DD47 pKa = 3.17MFEE50 pKa = 4.09PVEE53 pKa = 4.0
Molecular weight: 5.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6NYK6|A0A1M6NYK6_9CLOT Uncharacterized protein family (UPF0180) OS=Paramaledivibacter caminithermalis DSM 15212 OX=1121301 GN=SAMN02745912_01937 PE=4 SV=1
MM1 pKa = 7.74TYY3 pKa = 10.12QPKK6 pKa = 9.89KK7 pKa = 8.23RR8 pKa = 11.84QRR10 pKa = 11.84KK11 pKa = 8.39KK12 pKa = 8.49EE13 pKa = 3.5HH14 pKa = 6.11GFRR17 pKa = 11.84KK18 pKa = 9.77RR19 pKa = 11.84MKK21 pKa = 8.75TKK23 pKa = 10.24SGRR26 pKa = 11.84NVLKK30 pKa = 10.19RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 8.91GRR37 pKa = 11.84KK38 pKa = 9.02RR39 pKa = 11.84LTAA42 pKa = 4.18
MM1 pKa = 7.74TYY3 pKa = 10.12QPKK6 pKa = 9.89KK7 pKa = 8.23RR8 pKa = 11.84QRR10 pKa = 11.84KK11 pKa = 8.39KK12 pKa = 8.49EE13 pKa = 3.5HH14 pKa = 6.11GFRR17 pKa = 11.84KK18 pKa = 9.77RR19 pKa = 11.84MKK21 pKa = 8.75TKK23 pKa = 10.24SGRR26 pKa = 11.84NVLKK30 pKa = 10.19RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 8.91GRR37 pKa = 11.84KK38 pKa = 9.02RR39 pKa = 11.84LTAA42 pKa = 4.18
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1112298 |
39 |
2372 |
293.9 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.55 ± 0.041 | 1.059 ± 0.017 |
5.592 ± 0.031 | 7.434 ± 0.044 |
4.352 ± 0.031 | 6.42 ± 0.041 |
1.459 ± 0.015 | 10.65 ± 0.05 |
9.341 ± 0.046 | 9.11 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.018 | 5.964 ± 0.037 |
2.924 ± 0.024 | 2.314 ± 0.019 |
3.772 ± 0.027 | 5.814 ± 0.03 |
4.664 ± 0.024 | 6.152 ± 0.034 |
0.708 ± 0.013 | 4.125 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
