
Sedimentitalea nanhaiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Sedimentitalea
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
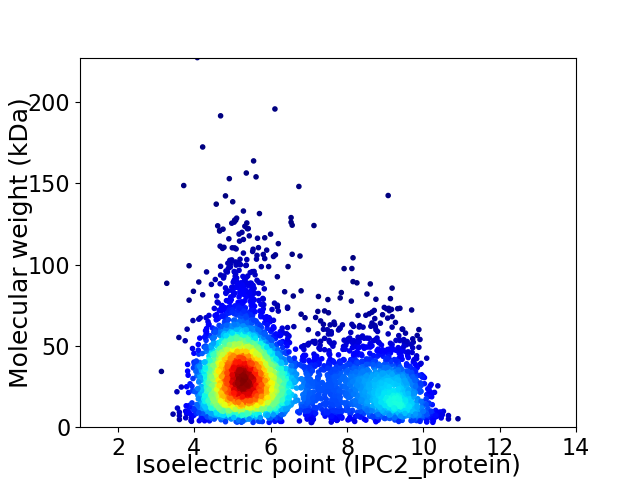
Virtual 2D-PAGE plot for 4762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6YJ17|A0A1I6YJ17_9RHOB NTE family protein OS=Sedimentitalea nanhaiensis OX=999627 GN=SAMN05216236_102244 PE=4 SV=1
MM1 pKa = 7.65SGIGNATRR9 pKa = 11.84YY10 pKa = 10.05VSASGDD16 pKa = 3.43AGIDD20 pKa = 3.88GILGDD25 pKa = 4.84RR26 pKa = 11.84AWDD29 pKa = 3.85DD30 pKa = 3.33AALLIGAPPTPDD42 pKa = 3.39VYY44 pKa = 11.33SGGYY48 pKa = 10.12GSGEE52 pKa = 4.21DD53 pKa = 3.74SGHH56 pKa = 5.2VQATAEE62 pKa = 4.34MTAMMQRR69 pKa = 11.84SLDD72 pKa = 3.73RR73 pKa = 11.84QAGSAADD80 pKa = 4.19DD81 pKa = 4.25GFSLEE86 pKa = 4.52GFTGQTVQQSTAASAHH102 pKa = 5.59IRR104 pKa = 11.84IAQTDD109 pKa = 4.26DD110 pKa = 4.02DD111 pKa = 4.56PFGYY115 pKa = 8.14GTAWAYY121 pKa = 10.53YY122 pKa = 8.72PSTGAASGDD131 pKa = 3.43VWFSTVSYY139 pKa = 10.56DD140 pKa = 3.15YY141 pKa = 10.04TYY143 pKa = 11.03PVAGNYY149 pKa = 10.41AYY151 pKa = 9.76VTMVHH156 pKa = 6.18EE157 pKa = 5.1LGHH160 pKa = 6.5ALGLEE165 pKa = 4.47HH166 pKa = 6.95GHH168 pKa = 6.33LTGSYY173 pKa = 8.25GTVPTSQDD181 pKa = 2.96SMEE184 pKa = 4.32YY185 pKa = 9.72TVMTYY190 pKa = 10.17RR191 pKa = 11.84SYY193 pKa = 11.03IGASPGNYY201 pKa = 9.77SNEE204 pKa = 3.74TWGYY208 pKa = 7.54AQSWMMLDD216 pKa = 3.66IAALQHH222 pKa = 5.93MYY224 pKa = 10.5GANFTTNSGDD234 pKa = 3.55TVYY237 pKa = 10.39KK238 pKa = 9.96WSPGSGATLVNGGIGIDD255 pKa = 3.49PGANRR260 pKa = 11.84IFATVWDD267 pKa = 4.31GGGTDD272 pKa = 3.75TFDD275 pKa = 3.4LSAYY279 pKa = 8.11ATALDD284 pKa = 3.69IDD286 pKa = 4.85LAPGAASVFDD296 pKa = 3.91SAQLARR302 pKa = 11.84LGPGEE307 pKa = 3.96YY308 pKa = 10.43ARR310 pKa = 11.84GNIYY314 pKa = 10.54NALLYY319 pKa = 10.82QGDD322 pKa = 3.79TRR324 pKa = 11.84SLIEE328 pKa = 4.27HH329 pKa = 6.23ATGGTGDD336 pKa = 4.55DD337 pKa = 3.59RR338 pKa = 11.84MSGNQADD345 pKa = 3.54NRR347 pKa = 11.84LRR349 pKa = 11.84GGDD352 pKa = 3.8GNDD355 pKa = 3.28TLMGQAGNDD364 pKa = 3.37TLMGQRR370 pKa = 11.84DD371 pKa = 4.11DD372 pKa = 5.73DD373 pKa = 3.82ILYY376 pKa = 10.97GNAGNDD382 pKa = 3.59TLKK385 pKa = 11.31GNGGNDD391 pKa = 3.26TLYY394 pKa = 11.41GNGDD398 pKa = 3.85DD399 pKa = 5.69DD400 pKa = 5.72LLIGHH405 pKa = 7.4GGHH408 pKa = 7.43DD409 pKa = 3.5SLYY412 pKa = 9.99GQDD415 pKa = 3.83GNDD418 pKa = 3.49VLRR421 pKa = 11.84GIAGQDD427 pKa = 3.29TLDD430 pKa = 4.32GGTGDD435 pKa = 5.66DD436 pKa = 3.59RR437 pKa = 11.84LFGNGGADD445 pKa = 3.54TLSGGAGQDD454 pKa = 3.48KK455 pKa = 10.88LFGAAGFDD463 pKa = 3.57MLYY466 pKa = 11.1GEE468 pKa = 5.43AGNDD472 pKa = 3.26QLTGGRR478 pKa = 11.84GADD481 pKa = 3.38QMTGGDD487 pKa = 3.62GADD490 pKa = 3.05VFRR493 pKa = 11.84FVAVNDD499 pKa = 3.94SPVGGGIDD507 pKa = 3.8TILDD511 pKa = 3.81FTPGSDD517 pKa = 4.42RR518 pKa = 11.84IDD520 pKa = 3.81LSGLAATPLGFQGFGNHH537 pKa = 6.17SGSAPDD543 pKa = 4.51VILQHH548 pKa = 6.19VSGDD552 pKa = 3.6TRR554 pKa = 11.84ILADD558 pKa = 4.43LDD560 pKa = 3.8GDD562 pKa = 3.96AQSDD566 pKa = 3.88LRR568 pKa = 11.84IDD570 pKa = 3.83LSGVLTLSGADD581 pKa = 3.96FIFF584 pKa = 4.49
MM1 pKa = 7.65SGIGNATRR9 pKa = 11.84YY10 pKa = 10.05VSASGDD16 pKa = 3.43AGIDD20 pKa = 3.88GILGDD25 pKa = 4.84RR26 pKa = 11.84AWDD29 pKa = 3.85DD30 pKa = 3.33AALLIGAPPTPDD42 pKa = 3.39VYY44 pKa = 11.33SGGYY48 pKa = 10.12GSGEE52 pKa = 4.21DD53 pKa = 3.74SGHH56 pKa = 5.2VQATAEE62 pKa = 4.34MTAMMQRR69 pKa = 11.84SLDD72 pKa = 3.73RR73 pKa = 11.84QAGSAADD80 pKa = 4.19DD81 pKa = 4.25GFSLEE86 pKa = 4.52GFTGQTVQQSTAASAHH102 pKa = 5.59IRR104 pKa = 11.84IAQTDD109 pKa = 4.26DD110 pKa = 4.02DD111 pKa = 4.56PFGYY115 pKa = 8.14GTAWAYY121 pKa = 10.53YY122 pKa = 8.72PSTGAASGDD131 pKa = 3.43VWFSTVSYY139 pKa = 10.56DD140 pKa = 3.15YY141 pKa = 10.04TYY143 pKa = 11.03PVAGNYY149 pKa = 10.41AYY151 pKa = 9.76VTMVHH156 pKa = 6.18EE157 pKa = 5.1LGHH160 pKa = 6.5ALGLEE165 pKa = 4.47HH166 pKa = 6.95GHH168 pKa = 6.33LTGSYY173 pKa = 8.25GTVPTSQDD181 pKa = 2.96SMEE184 pKa = 4.32YY185 pKa = 9.72TVMTYY190 pKa = 10.17RR191 pKa = 11.84SYY193 pKa = 11.03IGASPGNYY201 pKa = 9.77SNEE204 pKa = 3.74TWGYY208 pKa = 7.54AQSWMMLDD216 pKa = 3.66IAALQHH222 pKa = 5.93MYY224 pKa = 10.5GANFTTNSGDD234 pKa = 3.55TVYY237 pKa = 10.39KK238 pKa = 9.96WSPGSGATLVNGGIGIDD255 pKa = 3.49PGANRR260 pKa = 11.84IFATVWDD267 pKa = 4.31GGGTDD272 pKa = 3.75TFDD275 pKa = 3.4LSAYY279 pKa = 8.11ATALDD284 pKa = 3.69IDD286 pKa = 4.85LAPGAASVFDD296 pKa = 3.91SAQLARR302 pKa = 11.84LGPGEE307 pKa = 3.96YY308 pKa = 10.43ARR310 pKa = 11.84GNIYY314 pKa = 10.54NALLYY319 pKa = 10.82QGDD322 pKa = 3.79TRR324 pKa = 11.84SLIEE328 pKa = 4.27HH329 pKa = 6.23ATGGTGDD336 pKa = 4.55DD337 pKa = 3.59RR338 pKa = 11.84MSGNQADD345 pKa = 3.54NRR347 pKa = 11.84LRR349 pKa = 11.84GGDD352 pKa = 3.8GNDD355 pKa = 3.28TLMGQAGNDD364 pKa = 3.37TLMGQRR370 pKa = 11.84DD371 pKa = 4.11DD372 pKa = 5.73DD373 pKa = 3.82ILYY376 pKa = 10.97GNAGNDD382 pKa = 3.59TLKK385 pKa = 11.31GNGGNDD391 pKa = 3.26TLYY394 pKa = 11.41GNGDD398 pKa = 3.85DD399 pKa = 5.69DD400 pKa = 5.72LLIGHH405 pKa = 7.4GGHH408 pKa = 7.43DD409 pKa = 3.5SLYY412 pKa = 9.99GQDD415 pKa = 3.83GNDD418 pKa = 3.49VLRR421 pKa = 11.84GIAGQDD427 pKa = 3.29TLDD430 pKa = 4.32GGTGDD435 pKa = 5.66DD436 pKa = 3.59RR437 pKa = 11.84LFGNGGADD445 pKa = 3.54TLSGGAGQDD454 pKa = 3.48KK455 pKa = 10.88LFGAAGFDD463 pKa = 3.57MLYY466 pKa = 11.1GEE468 pKa = 5.43AGNDD472 pKa = 3.26QLTGGRR478 pKa = 11.84GADD481 pKa = 3.38QMTGGDD487 pKa = 3.62GADD490 pKa = 3.05VFRR493 pKa = 11.84FVAVNDD499 pKa = 3.94SPVGGGIDD507 pKa = 3.8TILDD511 pKa = 3.81FTPGSDD517 pKa = 4.42RR518 pKa = 11.84IDD520 pKa = 3.81LSGLAATPLGFQGFGNHH537 pKa = 6.17SGSAPDD543 pKa = 4.51VILQHH548 pKa = 6.19VSGDD552 pKa = 3.6TRR554 pKa = 11.84ILADD558 pKa = 4.43LDD560 pKa = 3.8GDD562 pKa = 3.96AQSDD566 pKa = 3.88LRR568 pKa = 11.84IDD570 pKa = 3.83LSGVLTLSGADD581 pKa = 3.96FIFF584 pKa = 4.49
Molecular weight: 60.33 kDa
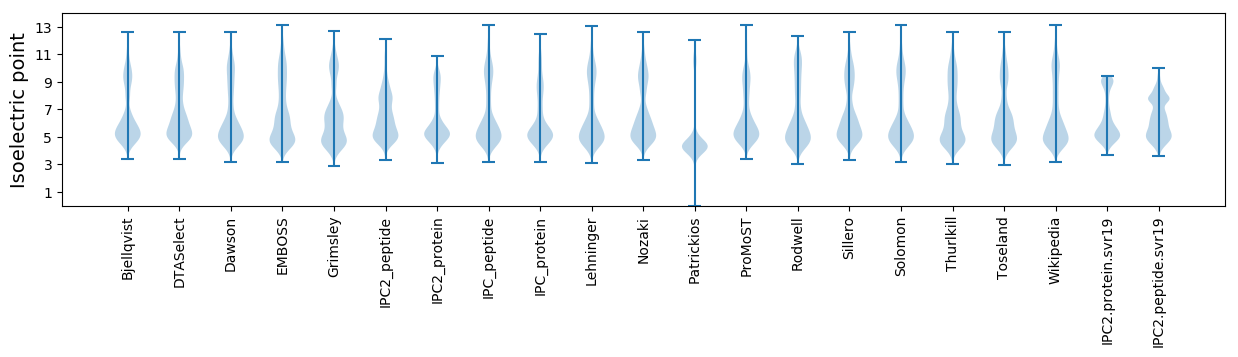
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7CMQ1|A0A1I7CMQ1_9RHOB Diguanylate cyclase/phosphodiesterase OS=Sedimentitalea nanhaiensis OX=999627 GN=SAMN05216236_1183 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1450164 |
28 |
2361 |
304.5 |
33.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.065 ± 0.049 | 0.938 ± 0.012 |
6.154 ± 0.031 | 5.448 ± 0.033 |
3.713 ± 0.024 | 8.634 ± 0.039 |
2.066 ± 0.018 | 5.26 ± 0.025 |
3.103 ± 0.03 | 10.03 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.839 ± 0.017 | 2.693 ± 0.02 |
5.104 ± 0.025 | 3.475 ± 0.021 |
6.835 ± 0.032 | 5.249 ± 0.028 |
5.515 ± 0.022 | 7.248 ± 0.029 |
1.385 ± 0.014 | 2.248 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
