
Reichenbachiella sp. 5M10
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Reichenbachiellaceae; Reichenbachiella; unclassified Reichenbachiella
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
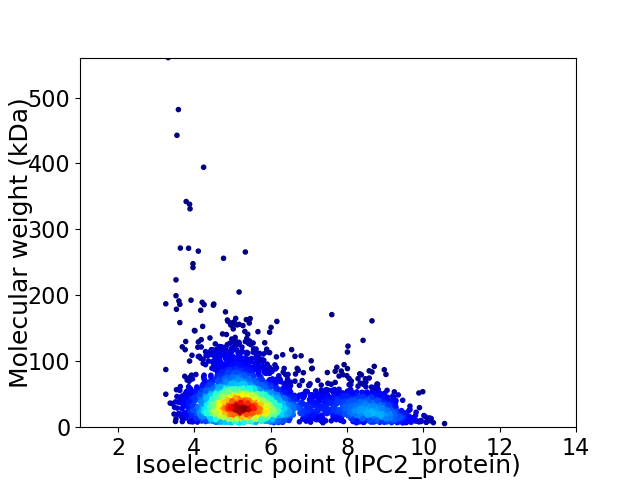
Virtual 2D-PAGE plot for 3696 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
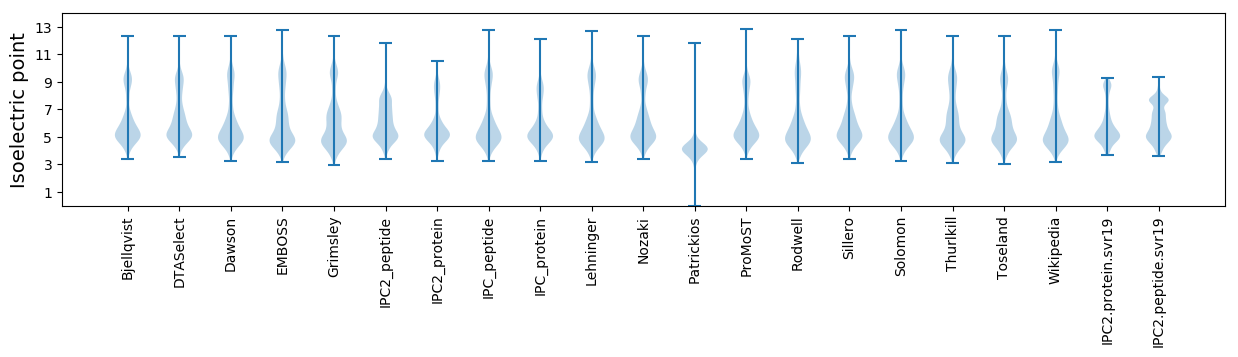
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G5L0P1|A0A2G5L0P1_9BACT Acyl-CoA oxidase OS=Reichenbachiella sp. 5M10 OX=1889772 GN=BFP72_03775 PE=3 SV=1
MM1 pKa = 7.39LAILSMVSVSYY12 pKa = 10.54AQNQAPVFTSTPVVTVDD29 pKa = 3.97DD30 pKa = 4.22NVAYY34 pKa = 10.43SYY36 pKa = 9.65TAYY39 pKa = 9.86TSDD42 pKa = 3.25SDD44 pKa = 3.65RR45 pKa = 11.84DD46 pKa = 3.81RR47 pKa = 11.84VTVTATTKK55 pKa = 10.51PSWLTLTNIDD65 pKa = 4.48AEE67 pKa = 4.61VTTFAGSGSSGSEE80 pKa = 3.97DD81 pKa = 3.2GHH83 pKa = 5.63GTSASFNNPSGIAIDD98 pKa = 3.7ASGNIYY104 pKa = 9.55VANANTIRR112 pKa = 11.84KK113 pKa = 8.78ISPSGDD119 pKa = 3.14VTTLAGSAVAGDD131 pKa = 3.6TDD133 pKa = 4.36GNGVNATFNLPKK145 pKa = 10.62GLAIDD150 pKa = 3.68AAGNVYY156 pKa = 10.46VADD159 pKa = 3.96TYY161 pKa = 11.13SFKK164 pKa = 10.32IRR166 pKa = 11.84KK167 pKa = 8.89ISTSGDD173 pKa = 3.21VTTLAGSGNVASIDD187 pKa = 3.72GNGANASFNYY197 pKa = 9.47PFALAVDD204 pKa = 3.93ATGNVYY210 pKa = 10.47VADD213 pKa = 4.47GGSQKK218 pKa = 9.59IRR220 pKa = 11.84KK221 pKa = 6.57ITPDD225 pKa = 3.44GEE227 pKa = 4.43VTTFAGSGDD236 pKa = 3.75NEE238 pKa = 4.14NVDD241 pKa = 4.2GNGLNASFISPRR253 pKa = 11.84GLTIDD258 pKa = 3.38AAGNLYY264 pKa = 8.33VTSFYY269 pKa = 11.13GHH271 pKa = 7.38AVRR274 pKa = 11.84KK275 pKa = 7.52ITPDD279 pKa = 3.34RR280 pKa = 11.84DD281 pKa = 3.44VTTLAGSGTAGNTDD295 pKa = 3.92DD296 pKa = 5.04NGTSASFNEE305 pKa = 4.21PAGIAVDD312 pKa = 4.24GSGNLYY318 pKa = 10.4VADD321 pKa = 3.88NRR323 pKa = 11.84NHH325 pKa = 7.48KK326 pKa = 9.21IRR328 pKa = 11.84KK329 pKa = 8.51ISTDD333 pKa = 3.06GDD335 pKa = 3.79VTTLAGSGNPGSTDD349 pKa = 3.28EE350 pKa = 4.88NGTSASFNNPIGVAVDD366 pKa = 3.49AAGSVYY372 pKa = 10.76VADD375 pKa = 3.96QVNHH379 pKa = 7.53KK380 pKa = 9.36IRR382 pKa = 11.84KK383 pKa = 6.73ISKK386 pKa = 7.95YY387 pKa = 10.13YY388 pKa = 9.72EE389 pKa = 4.06LSGSATDD396 pKa = 3.93QLGDD400 pKa = 3.75HH401 pKa = 6.48NVTLTASDD409 pKa = 3.96GKK411 pKa = 11.06GGTATQSFVLKK422 pKa = 9.57VAKK425 pKa = 10.17GRR427 pKa = 11.84AIGTGNILYY436 pKa = 10.03VDD438 pKa = 4.12KK439 pKa = 10.98NVSGGDD445 pKa = 3.46EE446 pKa = 4.59RR447 pKa = 11.84GSSWINAVPEE457 pKa = 4.08LADD460 pKa = 3.52ALVWAKK466 pKa = 11.15DD467 pKa = 3.58NYY469 pKa = 10.87DD470 pKa = 3.95DD471 pKa = 3.79TWATTPLKK479 pKa = 10.43IYY481 pKa = 8.86VAKK484 pKa = 9.34GTYY487 pKa = 9.09KK488 pKa = 10.6PLYY491 pKa = 10.01SPEE494 pKa = 5.55DD495 pKa = 3.67GDD497 pKa = 5.65NFGTDD502 pKa = 2.92QGRR505 pKa = 11.84NNSFSMVKK513 pKa = 10.08NVQLYY518 pKa = 11.07GGFDD522 pKa = 3.62PANGIEE528 pKa = 4.69DD529 pKa = 3.8LTDD532 pKa = 3.36TRR534 pKa = 11.84ILPSSDD540 pKa = 3.05EE541 pKa = 4.14AVGTILSGDD550 pKa = 3.27IGTADD555 pKa = 3.95DD556 pKa = 4.43ATDD559 pKa = 3.07NTYY562 pKa = 11.28NVLVSAGDD570 pKa = 3.42VGTASLDD577 pKa = 3.54GFSITDD583 pKa = 4.47GYY585 pKa = 11.06ANDD588 pKa = 3.69YY589 pKa = 10.91SDD591 pKa = 4.24IYY593 pKa = 11.72VNGKK597 pKa = 9.55QIFKK601 pKa = 10.56CSGAGVYY608 pKa = 10.6NSVSSPTYY616 pKa = 10.35KK617 pKa = 10.42HH618 pKa = 5.12VTLHH622 pKa = 6.27NNSCTEE628 pKa = 3.79SGGGMFSYY636 pKa = 10.05MSSPALQLVILKK648 pKa = 10.26NNQAKK653 pKa = 10.66DD654 pKa = 3.29GGGITNQEE662 pKa = 4.14SSSPTLLNVSIQSNNASEE680 pKa = 4.65NGSGMYY686 pKa = 10.25NVDD689 pKa = 3.21EE690 pKa = 4.84SEE692 pKa = 4.06PTLTNVSIVGNEE704 pKa = 3.62ASVSSEE710 pKa = 3.89EE711 pKa = 4.21VPSCVSQNSSLTLNNCIIWDD731 pKa = 4.22VVTGDD736 pKa = 3.48YY737 pKa = 8.23TAQNSLIKK745 pKa = 9.99GTSDD749 pKa = 3.07TSNGNLDD756 pKa = 3.63ATDD759 pKa = 4.07LTDD762 pKa = 3.36TDD764 pKa = 4.26IFTDD768 pKa = 4.19PANGDD773 pKa = 3.55YY774 pKa = 10.7SLKK777 pKa = 9.21STSIAVNSGSNTLYY791 pKa = 10.45TDD793 pKa = 3.87AGGDD797 pKa = 3.59LANDD801 pKa = 3.05VDD803 pKa = 3.74IAGNARR809 pKa = 11.84LYY811 pKa = 10.99DD812 pKa = 4.81GIPSTDD818 pKa = 3.69IIDD821 pKa = 3.66MGAYY825 pKa = 9.35EE826 pKa = 4.64YY827 pKa = 10.98LDD829 pKa = 4.51NIDD832 pKa = 4.29PVFTSTTTASFIEE845 pKa = 4.53NEE847 pKa = 4.04TGTAYY852 pKa = 9.71TVTATDD858 pKa = 4.21ANTITYY864 pKa = 10.55SLGSGNDD871 pKa = 3.0EE872 pKa = 4.04ALFDD876 pKa = 4.1IVGSTGVVTFKK887 pKa = 10.64AAPDD891 pKa = 3.85FEE893 pKa = 4.98SPADD897 pKa = 3.81ADD899 pKa = 3.65EE900 pKa = 4.93DD901 pKa = 3.67NAYY904 pKa = 10.02LINVIASDD912 pKa = 4.1GVNAANQNVTITVTDD927 pKa = 3.35VDD929 pKa = 3.92DD930 pKa = 4.3TDD932 pKa = 4.46PEE934 pKa = 4.5FTSATAVNFAEE945 pKa = 4.46NEE947 pKa = 3.96TGTAYY952 pKa = 9.71TVTATDD958 pKa = 4.16ANTVTYY964 pKa = 10.84SLGSGNDD971 pKa = 3.0EE972 pKa = 4.04ALFDD976 pKa = 4.11IVSSTGVVTFKK987 pKa = 10.02TAPDD991 pKa = 3.89FEE993 pKa = 5.23SPADD997 pKa = 3.7TDD999 pKa = 3.13EE1000 pKa = 4.83DD1001 pKa = 3.4NAYY1004 pKa = 9.74VINVIASDD1012 pKa = 3.91GVNTVNQNVTITVTDD1027 pKa = 3.31VDD1029 pKa = 3.78DD1030 pKa = 4.21TDD1032 pKa = 3.9PVFTSVTAVNFAEE1045 pKa = 4.42NEE1047 pKa = 3.96TGTAYY1052 pKa = 9.71TVTATDD1058 pKa = 4.16ANTVTYY1064 pKa = 10.84SLGSGNDD1071 pKa = 3.0EE1072 pKa = 4.04ALFDD1076 pKa = 4.1IVGSTGVVTFKK1087 pKa = 10.07TAPDD1091 pKa = 3.89FEE1093 pKa = 5.17NPADD1097 pKa = 4.06ADD1099 pKa = 3.66EE1100 pKa = 5.03DD1101 pKa = 3.69NAYY1104 pKa = 10.02LINVIASDD1112 pKa = 3.93GVNTVNQNVTITVTDD1127 pKa = 3.28VDD1129 pKa = 3.6DD1130 pKa = 4.24TEE1132 pKa = 4.92PVFSSTTAVNFAEE1145 pKa = 4.46NEE1147 pKa = 3.96TGTAYY1152 pKa = 9.71TVTATDD1158 pKa = 3.91ANTITYY1164 pKa = 10.17CLGSGNDD1171 pKa = 3.27EE1172 pKa = 4.73SIFNIISTSGVVTFKK1187 pKa = 10.71AAPDD1191 pKa = 3.87FEE1193 pKa = 5.43SPADD1197 pKa = 3.7TDD1199 pKa = 3.13EE1200 pKa = 4.83DD1201 pKa = 3.4NAYY1204 pKa = 9.74VINVIASDD1212 pKa = 3.91GVNTVNQNVTITVTDD1227 pKa = 3.35VDD1229 pKa = 3.75DD1230 pKa = 4.29TNPVFTSVTTASFIEE1245 pKa = 4.48NEE1247 pKa = 4.04TGTAYY1252 pKa = 9.71TVTATDD1258 pKa = 4.21ANTITYY1264 pKa = 10.55SLGSGNDD1271 pKa = 3.1EE1272 pKa = 4.7SIFNIISTSGVVTFKK1287 pKa = 10.04TAPDD1291 pKa = 3.89FEE1293 pKa = 5.23SPADD1297 pKa = 3.61TDD1299 pKa = 3.3EE1300 pKa = 5.48DD1301 pKa = 3.42NDD1303 pKa = 3.77YY1304 pKa = 10.71VINVIASDD1312 pKa = 3.6GGNTVNQNVTITVTDD1327 pKa = 3.31VDD1329 pKa = 3.78DD1330 pKa = 4.21TDD1332 pKa = 4.05PVFTSATTASFIEE1345 pKa = 4.5NEE1347 pKa = 4.04TGTAYY1352 pKa = 9.71TVTATDD1358 pKa = 4.21ANTITYY1364 pKa = 10.55SLGSGNDD1371 pKa = 3.0EE1372 pKa = 4.04ALFDD1376 pKa = 4.1IVGSTGVVTFKK1387 pKa = 10.0TAPNFEE1393 pKa = 4.68SPADD1397 pKa = 3.6TDD1399 pKa = 3.46ADD1401 pKa = 3.36NDD1403 pKa = 3.83YY1404 pKa = 10.43VINVIASDD1412 pKa = 3.91GVNTVNQNVTITVTDD1427 pKa = 3.33VDD1429 pKa = 4.3DD1430 pKa = 5.18VITATDD1436 pKa = 3.58SYY1438 pKa = 11.29EE1439 pKa = 3.89HH1440 pKa = 6.96ALNDD1444 pKa = 2.93ISLYY1448 pKa = 10.37PNPTHH1453 pKa = 7.05DD1454 pKa = 4.3QLNIDD1459 pKa = 4.31LSNYY1463 pKa = 9.04QNSNLSVRR1471 pKa = 11.84ITNVSGAQYY1480 pKa = 10.6FLRR1483 pKa = 11.84EE1484 pKa = 4.09NIATQRR1490 pKa = 11.84LLVNVEE1496 pKa = 4.4KK1497 pKa = 10.85YY1498 pKa = 10.06PSGVYY1503 pKa = 10.04LVILQTANHH1512 pKa = 5.93MVTKK1516 pKa = 10.27RR1517 pKa = 11.84VIIAPP1522 pKa = 3.91
MM1 pKa = 7.39LAILSMVSVSYY12 pKa = 10.54AQNQAPVFTSTPVVTVDD29 pKa = 3.97DD30 pKa = 4.22NVAYY34 pKa = 10.43SYY36 pKa = 9.65TAYY39 pKa = 9.86TSDD42 pKa = 3.25SDD44 pKa = 3.65RR45 pKa = 11.84DD46 pKa = 3.81RR47 pKa = 11.84VTVTATTKK55 pKa = 10.51PSWLTLTNIDD65 pKa = 4.48AEE67 pKa = 4.61VTTFAGSGSSGSEE80 pKa = 3.97DD81 pKa = 3.2GHH83 pKa = 5.63GTSASFNNPSGIAIDD98 pKa = 3.7ASGNIYY104 pKa = 9.55VANANTIRR112 pKa = 11.84KK113 pKa = 8.78ISPSGDD119 pKa = 3.14VTTLAGSAVAGDD131 pKa = 3.6TDD133 pKa = 4.36GNGVNATFNLPKK145 pKa = 10.62GLAIDD150 pKa = 3.68AAGNVYY156 pKa = 10.46VADD159 pKa = 3.96TYY161 pKa = 11.13SFKK164 pKa = 10.32IRR166 pKa = 11.84KK167 pKa = 8.89ISTSGDD173 pKa = 3.21VTTLAGSGNVASIDD187 pKa = 3.72GNGANASFNYY197 pKa = 9.47PFALAVDD204 pKa = 3.93ATGNVYY210 pKa = 10.47VADD213 pKa = 4.47GGSQKK218 pKa = 9.59IRR220 pKa = 11.84KK221 pKa = 6.57ITPDD225 pKa = 3.44GEE227 pKa = 4.43VTTFAGSGDD236 pKa = 3.75NEE238 pKa = 4.14NVDD241 pKa = 4.2GNGLNASFISPRR253 pKa = 11.84GLTIDD258 pKa = 3.38AAGNLYY264 pKa = 8.33VTSFYY269 pKa = 11.13GHH271 pKa = 7.38AVRR274 pKa = 11.84KK275 pKa = 7.52ITPDD279 pKa = 3.34RR280 pKa = 11.84DD281 pKa = 3.44VTTLAGSGTAGNTDD295 pKa = 3.92DD296 pKa = 5.04NGTSASFNEE305 pKa = 4.21PAGIAVDD312 pKa = 4.24GSGNLYY318 pKa = 10.4VADD321 pKa = 3.88NRR323 pKa = 11.84NHH325 pKa = 7.48KK326 pKa = 9.21IRR328 pKa = 11.84KK329 pKa = 8.51ISTDD333 pKa = 3.06GDD335 pKa = 3.79VTTLAGSGNPGSTDD349 pKa = 3.28EE350 pKa = 4.88NGTSASFNNPIGVAVDD366 pKa = 3.49AAGSVYY372 pKa = 10.76VADD375 pKa = 3.96QVNHH379 pKa = 7.53KK380 pKa = 9.36IRR382 pKa = 11.84KK383 pKa = 6.73ISKK386 pKa = 7.95YY387 pKa = 10.13YY388 pKa = 9.72EE389 pKa = 4.06LSGSATDD396 pKa = 3.93QLGDD400 pKa = 3.75HH401 pKa = 6.48NVTLTASDD409 pKa = 3.96GKK411 pKa = 11.06GGTATQSFVLKK422 pKa = 9.57VAKK425 pKa = 10.17GRR427 pKa = 11.84AIGTGNILYY436 pKa = 10.03VDD438 pKa = 4.12KK439 pKa = 10.98NVSGGDD445 pKa = 3.46EE446 pKa = 4.59RR447 pKa = 11.84GSSWINAVPEE457 pKa = 4.08LADD460 pKa = 3.52ALVWAKK466 pKa = 11.15DD467 pKa = 3.58NYY469 pKa = 10.87DD470 pKa = 3.95DD471 pKa = 3.79TWATTPLKK479 pKa = 10.43IYY481 pKa = 8.86VAKK484 pKa = 9.34GTYY487 pKa = 9.09KK488 pKa = 10.6PLYY491 pKa = 10.01SPEE494 pKa = 5.55DD495 pKa = 3.67GDD497 pKa = 5.65NFGTDD502 pKa = 2.92QGRR505 pKa = 11.84NNSFSMVKK513 pKa = 10.08NVQLYY518 pKa = 11.07GGFDD522 pKa = 3.62PANGIEE528 pKa = 4.69DD529 pKa = 3.8LTDD532 pKa = 3.36TRR534 pKa = 11.84ILPSSDD540 pKa = 3.05EE541 pKa = 4.14AVGTILSGDD550 pKa = 3.27IGTADD555 pKa = 3.95DD556 pKa = 4.43ATDD559 pKa = 3.07NTYY562 pKa = 11.28NVLVSAGDD570 pKa = 3.42VGTASLDD577 pKa = 3.54GFSITDD583 pKa = 4.47GYY585 pKa = 11.06ANDD588 pKa = 3.69YY589 pKa = 10.91SDD591 pKa = 4.24IYY593 pKa = 11.72VNGKK597 pKa = 9.55QIFKK601 pKa = 10.56CSGAGVYY608 pKa = 10.6NSVSSPTYY616 pKa = 10.35KK617 pKa = 10.42HH618 pKa = 5.12VTLHH622 pKa = 6.27NNSCTEE628 pKa = 3.79SGGGMFSYY636 pKa = 10.05MSSPALQLVILKK648 pKa = 10.26NNQAKK653 pKa = 10.66DD654 pKa = 3.29GGGITNQEE662 pKa = 4.14SSSPTLLNVSIQSNNASEE680 pKa = 4.65NGSGMYY686 pKa = 10.25NVDD689 pKa = 3.21EE690 pKa = 4.84SEE692 pKa = 4.06PTLTNVSIVGNEE704 pKa = 3.62ASVSSEE710 pKa = 3.89EE711 pKa = 4.21VPSCVSQNSSLTLNNCIIWDD731 pKa = 4.22VVTGDD736 pKa = 3.48YY737 pKa = 8.23TAQNSLIKK745 pKa = 9.99GTSDD749 pKa = 3.07TSNGNLDD756 pKa = 3.63ATDD759 pKa = 4.07LTDD762 pKa = 3.36TDD764 pKa = 4.26IFTDD768 pKa = 4.19PANGDD773 pKa = 3.55YY774 pKa = 10.7SLKK777 pKa = 9.21STSIAVNSGSNTLYY791 pKa = 10.45TDD793 pKa = 3.87AGGDD797 pKa = 3.59LANDD801 pKa = 3.05VDD803 pKa = 3.74IAGNARR809 pKa = 11.84LYY811 pKa = 10.99DD812 pKa = 4.81GIPSTDD818 pKa = 3.69IIDD821 pKa = 3.66MGAYY825 pKa = 9.35EE826 pKa = 4.64YY827 pKa = 10.98LDD829 pKa = 4.51NIDD832 pKa = 4.29PVFTSTTTASFIEE845 pKa = 4.53NEE847 pKa = 4.04TGTAYY852 pKa = 9.71TVTATDD858 pKa = 4.21ANTITYY864 pKa = 10.55SLGSGNDD871 pKa = 3.0EE872 pKa = 4.04ALFDD876 pKa = 4.1IVGSTGVVTFKK887 pKa = 10.64AAPDD891 pKa = 3.85FEE893 pKa = 4.98SPADD897 pKa = 3.81ADD899 pKa = 3.65EE900 pKa = 4.93DD901 pKa = 3.67NAYY904 pKa = 10.02LINVIASDD912 pKa = 4.1GVNAANQNVTITVTDD927 pKa = 3.35VDD929 pKa = 3.92DD930 pKa = 4.3TDD932 pKa = 4.46PEE934 pKa = 4.5FTSATAVNFAEE945 pKa = 4.46NEE947 pKa = 3.96TGTAYY952 pKa = 9.71TVTATDD958 pKa = 4.16ANTVTYY964 pKa = 10.84SLGSGNDD971 pKa = 3.0EE972 pKa = 4.04ALFDD976 pKa = 4.11IVSSTGVVTFKK987 pKa = 10.02TAPDD991 pKa = 3.89FEE993 pKa = 5.23SPADD997 pKa = 3.7TDD999 pKa = 3.13EE1000 pKa = 4.83DD1001 pKa = 3.4NAYY1004 pKa = 9.74VINVIASDD1012 pKa = 3.91GVNTVNQNVTITVTDD1027 pKa = 3.31VDD1029 pKa = 3.78DD1030 pKa = 4.21TDD1032 pKa = 3.9PVFTSVTAVNFAEE1045 pKa = 4.42NEE1047 pKa = 3.96TGTAYY1052 pKa = 9.71TVTATDD1058 pKa = 4.16ANTVTYY1064 pKa = 10.84SLGSGNDD1071 pKa = 3.0EE1072 pKa = 4.04ALFDD1076 pKa = 4.1IVGSTGVVTFKK1087 pKa = 10.07TAPDD1091 pKa = 3.89FEE1093 pKa = 5.17NPADD1097 pKa = 4.06ADD1099 pKa = 3.66EE1100 pKa = 5.03DD1101 pKa = 3.69NAYY1104 pKa = 10.02LINVIASDD1112 pKa = 3.93GVNTVNQNVTITVTDD1127 pKa = 3.28VDD1129 pKa = 3.6DD1130 pKa = 4.24TEE1132 pKa = 4.92PVFSSTTAVNFAEE1145 pKa = 4.46NEE1147 pKa = 3.96TGTAYY1152 pKa = 9.71TVTATDD1158 pKa = 3.91ANTITYY1164 pKa = 10.17CLGSGNDD1171 pKa = 3.27EE1172 pKa = 4.73SIFNIISTSGVVTFKK1187 pKa = 10.71AAPDD1191 pKa = 3.87FEE1193 pKa = 5.43SPADD1197 pKa = 3.7TDD1199 pKa = 3.13EE1200 pKa = 4.83DD1201 pKa = 3.4NAYY1204 pKa = 9.74VINVIASDD1212 pKa = 3.91GVNTVNQNVTITVTDD1227 pKa = 3.35VDD1229 pKa = 3.75DD1230 pKa = 4.29TNPVFTSVTTASFIEE1245 pKa = 4.48NEE1247 pKa = 4.04TGTAYY1252 pKa = 9.71TVTATDD1258 pKa = 4.21ANTITYY1264 pKa = 10.55SLGSGNDD1271 pKa = 3.1EE1272 pKa = 4.7SIFNIISTSGVVTFKK1287 pKa = 10.04TAPDD1291 pKa = 3.89FEE1293 pKa = 5.23SPADD1297 pKa = 3.61TDD1299 pKa = 3.3EE1300 pKa = 5.48DD1301 pKa = 3.42NDD1303 pKa = 3.77YY1304 pKa = 10.71VINVIASDD1312 pKa = 3.6GGNTVNQNVTITVTDD1327 pKa = 3.31VDD1329 pKa = 3.78DD1330 pKa = 4.21TDD1332 pKa = 4.05PVFTSATTASFIEE1345 pKa = 4.5NEE1347 pKa = 4.04TGTAYY1352 pKa = 9.71TVTATDD1358 pKa = 4.21ANTITYY1364 pKa = 10.55SLGSGNDD1371 pKa = 3.0EE1372 pKa = 4.04ALFDD1376 pKa = 4.1IVGSTGVVTFKK1387 pKa = 10.0TAPNFEE1393 pKa = 4.68SPADD1397 pKa = 3.6TDD1399 pKa = 3.46ADD1401 pKa = 3.36NDD1403 pKa = 3.83YY1404 pKa = 10.43VINVIASDD1412 pKa = 3.91GVNTVNQNVTITVTDD1427 pKa = 3.33VDD1429 pKa = 4.3DD1430 pKa = 5.18VITATDD1436 pKa = 3.58SYY1438 pKa = 11.29EE1439 pKa = 3.89HH1440 pKa = 6.96ALNDD1444 pKa = 2.93ISLYY1448 pKa = 10.37PNPTHH1453 pKa = 7.05DD1454 pKa = 4.3QLNIDD1459 pKa = 4.31LSNYY1463 pKa = 9.04QNSNLSVRR1471 pKa = 11.84ITNVSGAQYY1480 pKa = 10.6FLRR1483 pKa = 11.84EE1484 pKa = 4.09NIATQRR1490 pKa = 11.84LLVNVEE1496 pKa = 4.4KK1497 pKa = 10.85YY1498 pKa = 10.06PSGVYY1503 pKa = 10.04LVILQTANHH1512 pKa = 5.93MVTKK1516 pKa = 10.27RR1517 pKa = 11.84VIIAPP1522 pKa = 3.91
Molecular weight: 158.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G5L603|A0A2G5L603_9BACT Uncharacterized protein OS=Reichenbachiella sp. 5M10 OX=1889772 GN=BFP72_17490 PE=3 SV=1
MM1 pKa = 7.76AVRR4 pKa = 11.84KK5 pKa = 9.07LKK7 pKa = 10.43PITPGTRR14 pKa = 11.84YY15 pKa = 9.08RR16 pKa = 11.84VAPVFEE22 pKa = 5.67EE23 pKa = 4.18ITKK26 pKa = 8.26STPEE30 pKa = 3.6KK31 pKa = 10.52SLTKK35 pKa = 10.12GVHH38 pKa = 5.92RR39 pKa = 11.84NGGRR43 pKa = 11.84NDD45 pKa = 3.49SGKK48 pKa = 8.29MTIRR52 pKa = 11.84NVGGGHH58 pKa = 5.04KK59 pKa = 9.85RR60 pKa = 11.84KK61 pKa = 9.94YY62 pKa = 8.95RR63 pKa = 11.84TIDD66 pKa = 3.54FKK68 pKa = 10.96RR69 pKa = 11.84DD70 pKa = 2.99KK71 pKa = 10.67HH72 pKa = 6.35GIPAVVKK79 pKa = 10.39SIEE82 pKa = 4.04YY83 pKa = 10.84DD84 pKa = 3.28PMRR87 pKa = 11.84TARR90 pKa = 11.84IALLFYY96 pKa = 10.97ADD98 pKa = 3.49GEE100 pKa = 4.19KK101 pKa = 10.26RR102 pKa = 11.84YY103 pKa = 10.22IIAPHH108 pKa = 5.82GLKK111 pKa = 10.37VGQTVLSGSGVAPEE125 pKa = 4.42VGNTLPLSDD134 pKa = 4.77IPLGTIIHH142 pKa = 5.76NVEE145 pKa = 4.23LKK147 pKa = 9.92PGKK150 pKa = 9.21GACIARR156 pKa = 11.84SAGNYY161 pKa = 7.81VQLLARR167 pKa = 11.84EE168 pKa = 4.42GKK170 pKa = 9.36YY171 pKa = 9.1ATVKK175 pKa = 10.57LPSGEE180 pKa = 3.7MRR182 pKa = 11.84MILVVCLATIGTVSNGDD199 pKa = 3.28HH200 pKa = 6.22MNVVLGKK207 pKa = 10.26AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84SRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIFAKK253 pKa = 10.11GQKK256 pKa = 8.3TRR258 pKa = 11.84TPKK261 pKa = 10.31KK262 pKa = 9.23YY263 pKa = 9.95SNKK266 pKa = 10.06FIIGKK271 pKa = 9.41NKK273 pKa = 10.09
MM1 pKa = 7.76AVRR4 pKa = 11.84KK5 pKa = 9.07LKK7 pKa = 10.43PITPGTRR14 pKa = 11.84YY15 pKa = 9.08RR16 pKa = 11.84VAPVFEE22 pKa = 5.67EE23 pKa = 4.18ITKK26 pKa = 8.26STPEE30 pKa = 3.6KK31 pKa = 10.52SLTKK35 pKa = 10.12GVHH38 pKa = 5.92RR39 pKa = 11.84NGGRR43 pKa = 11.84NDD45 pKa = 3.49SGKK48 pKa = 8.29MTIRR52 pKa = 11.84NVGGGHH58 pKa = 5.04KK59 pKa = 9.85RR60 pKa = 11.84KK61 pKa = 9.94YY62 pKa = 8.95RR63 pKa = 11.84TIDD66 pKa = 3.54FKK68 pKa = 10.96RR69 pKa = 11.84DD70 pKa = 2.99KK71 pKa = 10.67HH72 pKa = 6.35GIPAVVKK79 pKa = 10.39SIEE82 pKa = 4.04YY83 pKa = 10.84DD84 pKa = 3.28PMRR87 pKa = 11.84TARR90 pKa = 11.84IALLFYY96 pKa = 10.97ADD98 pKa = 3.49GEE100 pKa = 4.19KK101 pKa = 10.26RR102 pKa = 11.84YY103 pKa = 10.22IIAPHH108 pKa = 5.82GLKK111 pKa = 10.37VGQTVLSGSGVAPEE125 pKa = 4.42VGNTLPLSDD134 pKa = 4.77IPLGTIIHH142 pKa = 5.76NVEE145 pKa = 4.23LKK147 pKa = 9.92PGKK150 pKa = 9.21GACIARR156 pKa = 11.84SAGNYY161 pKa = 7.81VQLLARR167 pKa = 11.84EE168 pKa = 4.42GKK170 pKa = 9.36YY171 pKa = 9.1ATVKK175 pKa = 10.57LPSGEE180 pKa = 3.7MRR182 pKa = 11.84MILVVCLATIGTVSNGDD199 pKa = 3.28HH200 pKa = 6.22MNVVLGKK207 pKa = 10.26AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84SRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIFAKK253 pKa = 10.11GQKK256 pKa = 8.3TRR258 pKa = 11.84TPKK261 pKa = 10.31KK262 pKa = 9.23YY263 pKa = 9.95SNKK266 pKa = 10.06FIIGKK271 pKa = 9.41NKK273 pKa = 10.09
Molecular weight: 29.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1385859 |
54 |
5295 |
375.0 |
42.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.934 ± 0.036 | 0.742 ± 0.012 |
6.079 ± 0.035 | 6.72 ± 0.043 |
4.602 ± 0.028 | 6.962 ± 0.046 |
2.056 ± 0.022 | 6.865 ± 0.037 |
5.987 ± 0.057 | 9.407 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.485 ± 0.019 | 4.892 ± 0.038 |
3.511 ± 0.019 | 3.98 ± 0.029 |
3.878 ± 0.032 | 6.886 ± 0.039 |
5.783 ± 0.063 | 6.774 ± 0.039 |
1.22 ± 0.018 | 4.236 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
